bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0534_orf3
Length=255
Score E
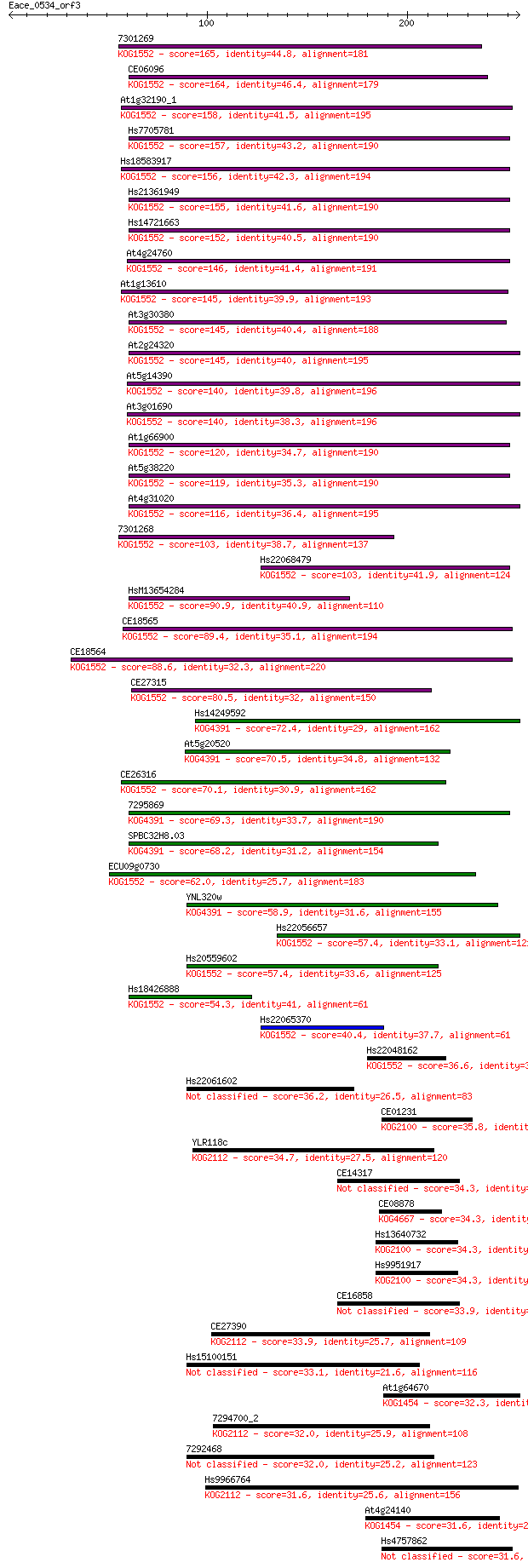
Sequences producing significant alignments: (Bits) Value
7301269 165 1e-40
CE06096 164 2e-40
At1g32190_1 158 1e-38
Hs7705781 157 2e-38
Hs18583917 156 4e-38
Hs21361949 155 6e-38
Hs14721663 152 6e-37
At4g24760 146 4e-35
At1g13610 145 7e-35
At3g30380 145 9e-35
At2g24320 145 1e-34
At5g14390 140 2e-33
At3g01690 140 3e-33
At1g66900 120 3e-27
At5g38220 119 6e-27
At4g31020 116 4e-26
7301268 103 3e-22
Hs22068479 103 5e-22
HsM13654284 90.9 2e-18
CE18565 89.4 7e-18
CE18564 88.6 1e-17
CE27315 80.5 3e-15
Hs14249592 72.4 8e-13
At5g20520 70.5 3e-12
CE26316 70.1 4e-12
7295869 69.3 8e-12
SPBC32H8.03 68.2 2e-11
ECU09g0730 62.0 1e-09
YNL320w 58.9 1e-08
Hs22056657 57.4 3e-08
Hs20559602 57.4 3e-08
Hs18426888 54.3 2e-07
Hs22065370 40.4 0.004
Hs22048162 36.6 0.058
Hs22061602 36.2 0.059
CE01231 35.8 0.087
YLR118c 34.7 0.19
CE14317 34.3 0.27
CE08878 34.3 0.27
Hs13640732 34.3 0.28
Hs9951917 34.3 0.28
CE16858 33.9 0.32
CE27390 33.9 0.34
Hs15100151 33.1 0.60
At1g64670 32.3 0.87
7294700_2 32.0 1.2
7292468 32.0 1.4
Hs9966764 31.6 1.7
At4g24140 31.6 1.7
Hs4757862 31.6 1.8
> 7301269
Length=286
Score = 165 bits (417), Expect = 1e-40, Method: Compositional matrix adjust.
Identities = 81/182 (44%), Positives = 115/182 (63%), Gaps = 1/182 (0%)
Query 56 RSGECTLLYSHGNAEDIFLSFSFFLHLSRVCRADVLLYEYVGYGCSSGKPSERGVYESVE 115
++ + TLL+SHGNA D+ SF+L L ++ Y+Y GYG S GKPSE+ +Y +E
Sbjct 87 KNAKYTLLFSHGNAVDLGQMSSFYLTLGSQINCNIFGYDYSGYGMSGGKPSEKNLYADIE 146
Query 116 AAYKYLTEELHIHPSKIVAYGRSLGSGPSVHLCASEEVGGLILQSALLSVHRVALR-LRL 174
AA++ + +I P I+ YG+S+G+ P+V L + EVG +IL S L+S RV R +
Sbjct 147 AAWQAMRTRFNISPETIILYGQSIGTVPTVDLASRHEVGAVILHSPLMSGLRVVFRNTKR 206
Query 175 TFPGDLFVNIKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNV 234
T+ D F +I K+ KVK PV IHG +DEV+ HGI +Y+R V P WV GAGHN+V
Sbjct 207 TWFFDAFPSIDKVAKVKAPVLVIHGTDDEVIDFSHGIGIYERCPKTVEPFWVEGAGHNDV 266
Query 235 EI 236
E+
Sbjct 267 EL 268
> CE06096
Length=405
Score = 164 bits (414), Expect = 2e-40, Method: Compositional matrix adjust.
Identities = 83/186 (44%), Positives = 114/186 (61%), Gaps = 13/186 (6%)
Query 61 TLLYSHGNAEDIFLSFSFFLHLSRVCRADVLLYEYVGYGCSSGKPSERGVYESVEAAYKY 120
TLL+SHGNA D+ SF L +V Y+Y GYGCS+GKPSE+ +Y + AA++
Sbjct 187 TLLFSHGNAVDLGQMTSFLYGLGFHLNCNVFSYDYSGYGCSTGKPSEKNLYADITAAFEL 246
Query 121 LTEELHIHPSKIVAYGRSLGSGPSVHLCASEEVGGLILQSALLSVHRVALRLRLTFPG-- 178
L E + KI+ YG+S+G+ PSV L + E++ L+L S L+S RVA FPG
Sbjct 247 LKSEFGVPKEKIILYGQSIGTVPSVDLASREDLAALVLHSPLMSGMRVA------FPGTT 300
Query 179 -----DLFVNIKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNN 233
D F +I+K+ +VKCP IHG +DEV+ HG+ +Y+R V PLWV GAGHN+
Sbjct 301 TTWCCDAFPSIEKVPRVKCPTLVIHGTDDEVIDFSHGVSIYERCPTSVEPLWVPGAGHND 360
Query 234 VEIVAG 239
VE+ A
Sbjct 361 VELHAA 366
> At1g32190_1
Length=289
Score = 158 bits (399), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 81/196 (41%), Positives = 120/196 (61%), Gaps = 3/196 (1%)
Query 57 SGECTLLYSHGNAEDIFLSFSFFLHLSRVCRADVLLYEYVGYGCSSGKPSERGVYESVEA 116
+ TLLYSHGNA D+ F F+ L R +++ Y+Y GYG S+GKPSE Y +EA
Sbjct 75 NARLTLLYSHGNAADLGQLFDLFVQLKVNLRVNLMGYDYSGYGASTGKPSEYDTYADIEA 134
Query 117 AYKYLTEELHIHPSKIVAYGRSLGSGPSVHLCAS-EEVGGLILQSALLSVHRVALRLRLT 175
AY+ L + + ++ YG+S+GSGP++HL + + G++L S +LS RV ++
Sbjct 135 AYECLQTDYGVGQEDLILYGQSVGSGPTLHLASKLPRLRGVVLHSGILSGLRVLCHVKFK 194
Query 176 FPGDLFVNIKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNVE 235
F D++ N+ KIKKVKCPV IHG D+VV HG L+K A+ PLW+ G GH N+E
Sbjct 195 FCCDIYSNVNKIKKVKCPVLVIHGTEDDVVNWLHGNRLWKMAKEPYEPLWIKGGGHCNLE 254
Query 236 IVAGREFLLAISRFLR 251
I +++ + RF++
Sbjct 255 IYP--DYIRHLYRFIQ 268
> Hs7705781
Length=293
Score = 157 bits (398), Expect = 2e-38, Method: Compositional matrix adjust.
Identities = 82/191 (42%), Positives = 115/191 (60%), Gaps = 3/191 (1%)
Query 61 TLLYSHGNAEDIFLSFSFFLHLSRVCRADVLLYEYVGYGCSSGKPSERGVYESVEAAYKY 120
TLL+SHGNA D+ SF++ L ++ Y+Y GYG SSGKP+E+ +Y +EAA+
Sbjct 93 TLLFSHGNAVDLGQMSSFYIGLGSRINCNIFSYDYSGYGASSGKPTEKNLYADIEAAWLA 152
Query 121 LTEELHIHPSKIVAYGRSLGSGPSVHLCASEEVGGLILQSALLSVHRVAL-RLRLTFPGD 179
L I P ++ YG+S+G+ PSV L A E +IL S L S RVA + T+ D
Sbjct 153 LRTRYGIRPENVIIYGQSIGTVPSVDLAARYESAAVILHSPLTSGMRVAFPDTKETYCFD 212
Query 180 LFVNIKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNVEIVAG 239
F NI KI K+ PV IHG DEV+ HG+ L++R + V PLWV GAGHN+VE+
Sbjct 213 AFPNIDKISKITSPVLIIHGTEDEVIDFSHGLALFERCQRPVEPLWVEGAGHNDVELYG- 271
Query 240 REFLLAISRFL 250
++L + +F+
Sbjct 272 -QYLERLKQFV 281
> Hs18583917
Length=329
Score = 156 bits (394), Expect = 4e-38, Method: Compositional matrix adjust.
Identities = 82/195 (42%), Positives = 118/195 (60%), Gaps = 3/195 (1%)
Query 57 SGECTLLYSHGNAEDIFLSFSFFLHLSRVCRADVLLYEYVGYGCSSGKPSERGVYESVEA 116
S TLL+SHGNA D+ SF++ L ++ Y+Y GYG SSGKPSE+ +Y ++A
Sbjct 130 SSRYTLLFSHGNAVDLGQMCSFYIGLGSRINCNIFSYDYSGYGVSSGKPSEKNLYADIDA 189
Query 117 AYKYLTEELHIHPSKIVAYGRSLGSGPSVHLCASEEVGGLILQSALLSVHRVAL-RLRLT 175
A++ L + P I+ YG+S+G+ P+V L + E +IL S L+S RVA R T
Sbjct 190 AWQALRTRYGVSPENIILYGQSIGTVPTVDLASRYECAAVILHSPLMSGLRVAFPDTRKT 249
Query 176 FPGDLFVNIKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNVE 235
+ D F +I KI KV PV IHG DEV+ HG+ +Y+R V PLWV GAGHN++E
Sbjct 250 YCFDAFPSIDKISKVTSPVLVIHGTEDEVIDFSHGLAMYERCPRAVEPLWVEGAGHNDIE 309
Query 236 IVAGREFLLAISRFL 250
+ A ++L + +F+
Sbjct 310 LYA--QYLERLKQFI 322
> Hs21361949
Length=361
Score = 155 bits (393), Expect = 6e-38, Method: Compositional matrix adjust.
Identities = 79/191 (41%), Positives = 117/191 (61%), Gaps = 3/191 (1%)
Query 61 TLLYSHGNAEDIFLSFSFFLHLSRVCRADVLLYEYVGYGCSSGKPSERGVYESVEAAYKY 120
T+L+SHGNA D+ SF++ L ++ Y+Y GYG SSG+PSER +Y ++AA++
Sbjct 164 TVLFSHGNAVDLGQMSSFYIGLGSRLHCNIFSYDYSGYGASSGRPSERNLYADIDAAWQA 223
Query 121 LTEELHIHPSKIVAYGRSLGSGPSVHLCASEEVGGLILQSALLSVHRVAL-RLRLTFPGD 179
L I P I+ YG+S+G+ P+V L + E ++L S L S RVA + T+ D
Sbjct 224 LRTRYGISPDSIILYGQSIGTVPTVDLASRYECAAVVLHSPLTSGMRVAFPDTKKTYCFD 283
Query 180 LFVNIKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNVEIVAG 239
F NI+K+ K+ PV IHG DEV+ HG+ LY+R V PLWV GAGHN++E+ +
Sbjct 284 AFPNIEKVSKITSPVLIIHGTEDEVIDFSHGLALYERCPKAVEPLWVEGAGHNDIELYS- 342
Query 240 REFLLAISRFL 250
++L + RF+
Sbjct 343 -QYLERLRRFI 352
> Hs14721663
Length=310
Score = 152 bits (384), Expect = 6e-37, Method: Compositional matrix adjust.
Identities = 77/191 (40%), Positives = 116/191 (60%), Gaps = 3/191 (1%)
Query 61 TLLYSHGNAEDIFLSFSFFLHLSRVCRADVLLYEYVGYGCSSGKPSERGVYESVEAAYKY 120
T+L+SHGNA D+ SF++ L ++ Y+ GYG SSG+PSER +Y ++A ++
Sbjct 113 TVLFSHGNAVDLGQMSSFYIGLGSRLHCNIFTYDSSGYGASSGRPSERNLYADIDATWQA 172
Query 121 LTEELHIHPSKIVAYGRSLGSGPSVHLCASEEVGGLILQSALLSVHRVALR-LRLTFPGD 179
L I P I+ YG+S+G+ P++ L + E ++L S L S RVA R + T+ D
Sbjct 173 LRTRYGISPDSIILYGQSIGTVPTMDLASRYECAAVVLHSPLTSGMRVAFRDTKKTYCFD 232
Query 180 LFVNIKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNVEIVAG 239
F NI+K+ K+ PV IHG DEV+ HG+ LY+R V PLWV GAGHN++E+ +
Sbjct 233 AFPNIEKVSKITSPVLIIHGREDEVIDFSHGLALYERCPKAVEPLWVEGAGHNDIELYS- 291
Query 240 REFLLAISRFL 250
++L + RF+
Sbjct 292 -QYLERLRRFI 301
> At4g24760
Length=365
Score = 146 bits (368), Expect = 4e-35, Method: Compositional matrix adjust.
Identities = 79/192 (41%), Positives = 115/192 (59%), Gaps = 3/192 (1%)
Query 60 CTLLYSHGNAEDIFLSFSFFLHLSRVCRADVLLYEYVGYGCSSGKPSERGVYESVEAAYK 119
TLLYSHGNA DI + F+ LS R +++ Y+Y GYG SSGKP+E+ Y +EAAYK
Sbjct 69 TTLLYSHGNAADIGQMYELFIELSIHLRVNLMGYDYSGYGQSSGKPTEQNTYADIEAAYK 128
Query 120 YLTEELHIHPSKIVAYGRSLGSGPSVHLCAS-EEVGGLILQSALLSVHRVALRLRLTFPG 178
L E I+ YG+S+GSGP+V L A + IL S +LS RV ++ T+
Sbjct 129 CLEENYGAKQENIILYGQSVGSGPTVDLAARLPRLRASILHSPILSGLRVMYPVKRTYWF 188
Query 179 DLFVNIKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNVEIVA 238
D++ NI KI V+CPV IHG D+VV HG +L++ + + PLW+ G H ++E+
Sbjct 189 DIYKNIDKITLVRCPVLVIHGTADDVVDFSHGKQLWELCQEKYEPLWLKGGNHCDLELFP 248
Query 239 GREFLLAISRFL 250
E++ + +F+
Sbjct 249 --EYIGHLKKFV 258
> At1g13610
Length=351
Score = 145 bits (367), Expect = 7e-35, Method: Compositional matrix adjust.
Identities = 77/199 (38%), Positives = 119/199 (59%), Gaps = 7/199 (3%)
Query 57 SGECTLLYSHGNAEDIFLSFSFFLHLSRVCRADVLLYEYVGYGCSSGKPSERGVYESVEA 116
+ + T+L+SHGNA D+ F L ++ +++ Y+Y GYG SSGKPSE+ Y +EA
Sbjct 63 TAKLTVLFSHGNASDLAQIFYILAELIQL-NVNLMGYDYSGYGQSSGKPSEQDTYADIEA 121
Query 117 AYKYLTEELHIHPSKIVAYGRSLGSGPSVHLCAS-EEVGGLILQSALLSVHRVALRLRLT 175
AY +L + +I+ YG+S+GSGPS+ L + + L+L S LS RV ++ +
Sbjct 122 AYNWLRQTYGTKDERIILYGQSVGSGPSLELASRLPRLRALVLHSPFLSGLRVMYPVKHS 181
Query 176 FPGDLFVNIKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNVE 235
FP D++ NI KI V+CPV IHG +D+VV I HG L+ + + PLW+ G GH+++E
Sbjct 182 FPFDIYKNIDKIHLVECPVLVIHGTDDDVVNISHGKHLWGLCKEKYEPLWLKGRGHSDIE 241
Query 236 IVAG-----REFLLAISRF 249
+ R+F+ AI +
Sbjct 242 MSPEYLPHLRKFISAIEKL 260
> At3g30380
Length=399
Score = 145 bits (366), Expect = 9e-35, Method: Compositional matrix adjust.
Identities = 76/194 (39%), Positives = 118/194 (60%), Gaps = 6/194 (3%)
Query 61 TLLYSHGNAEDIFLSFSFFLHLSRVCRADVLLYEYVGYGCSSGKPSERGVYESVEAAYKY 120
TLLYSHGNA D+ F F LS R +++ Y+Y GYG SSGKPSE+ Y +EA Y+
Sbjct 69 TLLYSHGNAADLGQMFELFSELSLHLRVNLIGYDYSGYGRSSGKPSEQNTYSDIEAVYRC 128
Query 121 LTEELHIHPSKIVAYGRSLGSGPSVHLCAS-EEVGGLILQSALLSVHRVALRLRLTFPGD 179
L E+ + ++ YG+S+GSGP++ L + + ++L SA+ S RV ++ T+ D
Sbjct 129 LEEKYGVKEQDVILYGQSVGSGPTLELASRLPNLRAVVLHSAIASGLRVMYPVKRTYWFD 188
Query 180 LFVNIKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNVEIVAG 239
++ N++KI VKCPV IHG +D+VV HG +L++ + + PLW+ G H ++E+
Sbjct 189 IYKNVEKISFVKCPVLVIHGTSDDVVNWSHGKQLFELCKEKYEPLWIKGGNHCDLELYPQ 248
Query 240 -----REFLLAISR 248
R+F+ AI +
Sbjct 249 YIKHLRKFVSAIEK 262
> At2g24320
Length=316
Score = 145 bits (365), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 78/219 (35%), Positives = 121/219 (55%), Gaps = 26/219 (11%)
Query 61 TLLYSHGNAEDIFLSFSFFLHLSRVCRADVLLYEYVGYGCSSGKPSERGVYESVEAAYKY 120
TLLYSHGNA D+ F+ L R +++ Y+Y GYG S+GKP+E Y +EA Y
Sbjct 69 TLLYSHGNAADLGQMVDLFIELRAHLRVNIMSYDYSGYGASTGKPTELNTYYDIEAVYNC 128
Query 121 LTEELHIHPSKIVAYGRSLGSGPSVHLCAS-EEVGGLILQSALLSVHRVALRLRLTFPGD 179
L E I +++ YG+S+GSGP++HL + + + G++L SA+LS RV +++TF D
Sbjct 129 LRTEYGIMQEEMILYGQSVGSGPTLHLASRVKRLRGIVLHSAILSGLRVLYPVKMTFWFD 188
Query 180 LF-----------------------VNIKKIKKVKCPVFCIHGANDEVVPIHHGIELYKR 216
++ NI KI+ V CPV IHG D++V + HG L++
Sbjct 189 MYKVSLISLVSGYYYRVSLSNSGILQNIDKIRHVTCPVLVIHGTKDDIVNMSHGKRLWEL 248
Query 217 ARVRVSPLWVGGAGHNNVEIVAGREFLLAISRFLRLLLR 255
A+ + PLWV G GH N+E E++ + +F+ + +
Sbjct 249 AKDKYDPLWVKGGGHCNLETYP--EYIKHMRKFMNAMEK 285
> At5g14390
Length=369
Score = 140 bits (354), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 78/197 (39%), Positives = 116/197 (58%), Gaps = 3/197 (1%)
Query 60 CTLLYSHGNAEDIFLSFSFFLHLSRVCRADVLLYEYVGYGCSSGKPSERGVYESVEAAYK 119
TLLYSHGNA D+ + F+ LS + +++ Y+Y GYG S+GKPSE Y +EAAYK
Sbjct 69 STLLYSHGNAADLGQMYELFIELSIHLKVNLMGYDYSGYGQSTGKPSEHHTYADIEAAYK 128
Query 120 YLTEELHIHPSKIVAYGRSLGSGPSVHLCAS-EEVGGLILQSALLSVHRVALRLRLTFPG 178
L E I+ YG+S+GSGP++ L A ++ +L S +LS RV ++ T+
Sbjct 129 CLEETYGAKQEDIILYGQSVGSGPTLDLAARLPQLRAAVLHSPILSGLRVMYPVKKTYWF 188
Query 179 DLFVNIKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNVEIVA 238
D+F NI KI V CPV IHG DEVV HG +L++ ++ + PLW+ G H ++E
Sbjct 189 DIFKNIDKIPLVNCPVLVIHGTCDEVVDCSHGKQLWELSKEKYEPLWLEGGNHCDLEHYP 248
Query 239 GREFLLAISRFLRLLLR 255
E++ + +F+ + R
Sbjct 249 --EYIKHLKKFITTVER 263
> At3g01690
Length=361
Score = 140 bits (352), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 75/197 (38%), Positives = 116/197 (58%), Gaps = 3/197 (1%)
Query 60 CTLLYSHGNAEDIFLSFSFFLHLSRVCRADVLLYEYVGYGCSSGKPSERGVYESVEAAYK 119
TLLYSHGNA D+ + F+ LS + +++ Y+Y GYG S+GKPSE Y +EA YK
Sbjct 69 STLLYSHGNAADLGQMYELFIELSIHLKVNLMGYDYSGYGQSTGKPSEHNTYADIEAVYK 128
Query 120 YLTEELHIHPSKIVAYGRSLGSGPSVHLCAS-EEVGGLILQSALLSVHRVALRLRLTFPG 178
L E ++ YG+S+GSGP++ L + ++ ++L S +LS RV ++ T+
Sbjct 129 CLEETFGSKQEGVILYGQSVGSGPTLDLASRLPQLRAVVLHSPILSGLRVMYSVKKTYWF 188
Query 179 DLFVNIKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNVEIVA 238
D++ NI KI V CPV IHG +DEVV HG +L++ + + PLWV G H ++E
Sbjct 189 DIYKNIDKIPYVDCPVLIIHGTSDEVVDCSHGKQLWELCKDKYEPLWVKGGNHCDLEHYP 248
Query 239 GREFLLAISRFLRLLLR 255
E++ + +F+ + R
Sbjct 249 --EYIRHLKKFIATVER 263
> At1g66900
Length=256
Score = 120 bits (300), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 66/191 (34%), Positives = 109/191 (57%), Gaps = 19/191 (9%)
Query 61 TLLYSHGNAEDIFLSFSFFLHLSRVCRADVLLYEYVGYGCSSGKPSERGVYESVEAAYKY 120
TLLYSHGNA D+ F F+ LS R +++ Y+Y GYG S+G+ SE Y +EA+YK
Sbjct 71 TLLYSHGNAADLGQMFELFVELSNRLRVNLMGYDYSGYGQSTGQASECNTYADIEASYKC 130
Query 121 LTEELHIHPSKIVAYGRSLGSGPSVHLCA-SEEVGGLILQSALLSVHRVALRLRLTFPGD 179
L E+ + +++ YG+S+GSGP+V L + + + G++LQ +LS RV ++ T+ D
Sbjct 131 LKEKYGVKDDQLIVYGQSVGSGPTVDLASRTPNLRGVVLQCPILSGMRVLYPVKCTYWFD 190
Query 180 LFVNIKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNVEIVAG 239
++ G DEVV HG L++ ++ + PLW+ G GH ++E+
Sbjct 191 IY----------------KGTADEVVDWSHGKRLWELSKEKYEPLWISGGGHCDLELYP- 233
Query 240 REFLLAISRFL 250
+F+ + +F+
Sbjct 234 -DFIRHLKKFV 243
> At5g38220
Length=320
Score = 119 bits (298), Expect = 6e-27, Method: Compositional matrix adjust.
Identities = 67/191 (35%), Positives = 108/191 (56%), Gaps = 19/191 (9%)
Query 61 TLLYSHGNAEDIFLSFSFFLHLSRVCRADVLLYEYVGYGCSSGKPSERGVYESVEAAYKY 120
TLLYSHGNA D+ F F+ LS R +++ Y+Y GYG S+GK SE Y ++AAY
Sbjct 68 TLLYSHGNAADLGQMFELFIELSNRLRLNLMGYDYSGYGQSTGKASECNTYADIDAAYTC 127
Query 121 LTEELHIHPSKIVAYGRSLGSGPSVHLCA-SEEVGGLILQSALLSVHRVALRLRLTFPGD 179
L E + +++ YG+S+GSGP++ L + + + G++L S +LS RV ++ T+ D
Sbjct 128 LKEHYGVKDDQLILYGQSVGSGPTIDLASRTPNLRGVVLHSPILSGMRVLYPVKRTYWFD 187
Query 180 LFVNIKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNVEIVAG 239
++ G DEVV HG +L++ ++ + PLWV G GH N+E+
Sbjct 188 IY----------------KGTADEVVDCSHGKQLWELSKEKYEPLWVSGGGHCNLELYP- 230
Query 240 REFLLAISRFL 250
EF+ + +++
Sbjct 231 -EFIKHLKKYV 240
> At4g31020
Length=307
Score = 116 bits (291), Expect = 4e-26, Method: Compositional matrix adjust.
Identities = 71/214 (33%), Positives = 111/214 (51%), Gaps = 26/214 (12%)
Query 61 TLLYSHGNAEDIFLSFSFFLHLSRVCRADVLLYEYVGYGCSSGKPSERGVYESVEAAYKY 120
TLLYSHGNA D+ F+ L R +++ Y + PSE Y +EA Y
Sbjct 70 TLLYSHGNAADLGQMVELFIELRAHLRVNIMRYI-----LKTLMPSEFNTYYDIEAVYSC 124
Query 121 LTEELHIHPSKIVAYGRSLGSGPSVHLCAS-EEVGGLILQSALLSVHRVALRLRLTFPGD 179
L + I +I+ YG+S+GSGP++H+ + + + G++L SA+LS RV +++T D
Sbjct 125 LRSDYGIKQEEIILYGQSVGSGPTLHMASRLKRLRGVVLHSAILSGIRVLYPVKMTLWFD 184
Query 180 LF------------------VNIKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRV 221
+F NI KI+ V V IHG NDE+V + HG L++ A+ +
Sbjct 185 IFKVRKAHTKDLLLVGLHIYSNIDKIRHVNSQVLVIHGTNDEIVDLSHGKRLWELAKEKY 244
Query 222 SPLWVGGAGHNNVEIVAGREFLLAISRFLRLLLR 255
PLWV G GH N+E E++ + +F+ + +
Sbjct 245 DPLWVKGGGHCNLETYP--EYIKHLKKFVNAMEK 276
> 7301268
Length=244
Score = 103 bits (258), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 53/138 (38%), Positives = 81/138 (58%), Gaps = 1/138 (0%)
Query 56 RSGECTLLYSHGNAEDIFLSFSFFLHLSRVCRADVLLYEYVGYGCSSGKPSERGVYESVE 115
++ + TLL+SHGNA D+ SF+L L ++ Y+Y GYG S GKPSE+ +Y +E
Sbjct 87 KNAKYTLLFSHGNAVDLGQMSSFYLTLGSQINCNIFGYDYSGYGMSGGKPSEKNLYADIE 146
Query 116 AAYKYLTEELHIHPSKIVAYGRSLGSGPSVHLCASEEVGGLILQSALLSVHRVALR-LRL 174
AA++ + +I P I+ YG+S+G+ P+V L + EVG +IL S L+S RV R +
Sbjct 147 AAWQAMRTRFNISPETIILYGQSIGTVPTVDLASRHEVGAVILHSPLMSGLRVVFRNTKR 206
Query 175 TFPGDLFVNIKKIKKVKC 192
T+ D + + I C
Sbjct 207 TWFFDAYFRVLDIMMWNC 224
> Hs22068479
Length=161
Score = 103 bits (256), Expect = 5e-22, Method: Compositional matrix adjust.
Identities = 52/124 (41%), Positives = 75/124 (60%), Gaps = 2/124 (1%)
Query 127 IHPSKIVAYGRSLGSGPSVHLCASEEVGGLILQSALLSVHRVALRLRLTFPGDLFVNIKK 186
I P I+ YG+S+G+ P+V L + E ++L S L S RVA T+ D F NI+K
Sbjct 31 ISPDSIILYGQSIGTVPTVDLASRYECAAVVLHSPLTSGMRVAFPDTKTYCFDAFPNIEK 90
Query 187 IKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNVEIVAGREFLLAI 246
+ K+ PV IHG DEV+ HG+ LY+R V PLWV GAGHN++E+ + ++L +
Sbjct 91 VSKITSPVLIIHGMEDEVIDFSHGLALYERCPKAVEPLWVEGAGHNDIELYS--QYLERL 148
Query 247 SRFL 250
RF+
Sbjct 149 RRFI 152
> HsM13654284
Length=236
Score = 90.9 bits (224), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 45/110 (40%), Positives = 67/110 (60%), Gaps = 0/110 (0%)
Query 61 TLLYSHGNAEDIFLSFSFFLHLSRVCRADVLLYEYVGYGCSSGKPSERGVYESVEAAYKY 120
T+L+SHGNA D+ SF++ L ++ Y+Y GYG SSG+PSER +Y ++AA++
Sbjct 113 TVLFSHGNAVDLGQMSSFYIGLGSRLHCNIFSYDYSGYGASSGRPSERNLYADIDAAWQA 172
Query 121 LTEELHIHPSKIVAYGRSLGSGPSVHLCASEEVGGLILQSALLSVHRVAL 170
L I P I+ YG+S+G+ P+V L + E ++L S L S RVA
Sbjct 173 LRTRYGISPDSIILYGQSIGTVPTVDLASRYECAAVVLHSPLTSGMRVAF 222
> CE18565
Length=305
Score = 89.4 bits (220), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 68/210 (32%), Positives = 105/210 (50%), Gaps = 25/210 (11%)
Query 58 GECTLLYSHGNAEDI--FLSFS--FFLHLSRVCRADVLLYEYVGYGCSSGKPSERGVYES 113
E +L+ N+ D+ FL + F+ + V D+ ++Y GYG SSG E+ VY
Sbjct 76 AEQVVLFCQPNSSDLGGFLQPNSMNFVTYANVFETDLYAFDYSGYGFSSGTQGEKNVYAD 135
Query 114 VEAAYKYLTEELHIHPSK-IVAYGRSLGSGPSVHLCAS--EEVGGLILQSALLSVHRVAL 170
V A Y+ + L + P K IV G S+G+ +V L A+ + + G++L + S L
Sbjct 136 VRAVYEKI---LEMRPDKKIVVMGYSIGTTAAVDLAATNPDRLAGVVLIAPFTS----GL 188
Query 171 RLRLTFP-------GDLFVNIKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSP 223
RL + P D F + KI + V HG DEV+P+ HG+ LY++ + V P
Sbjct 189 RLFSSKPDKPDTCWADSFKSFDKINNIDTRVLICHGDVDEVIPLSHGLALYEKLKNPVPP 248
Query 224 LWVGGAGHNNVEIVAGR--EFLLAISRFLR 251
L V GA H+ I++G+ I+ FLR
Sbjct 249 LIVHGANHHT--ILSGKYIHVFTRIANFLR 276
> CE18564
Length=335
Score = 88.6 bits (218), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 71/225 (31%), Positives = 111/225 (49%), Gaps = 21/225 (9%)
Query 32 CCCCYPSAAAATLLLLLLLLLFVGRSGECTLLYSHGNAEDIFLSFSFFLHLSRVCRADVL 91
C C P+ + + ++LF S + N+ ++FS F +L DV
Sbjct 104 CVKCSPNCYSKNPEVANQVVLFCQSSSADLGSFLQPNS----MNFSTFANL---FETDVY 156
Query 92 LYEYVGYGCSSGKPSERGVYESVEAAYKYLTEELHIHPSK-IVAYGRSLGSGPSVHLCAS 150
++Y GYG SSG SE+ +Y V A Y+++ L P K IV G S+G+ +V L AS
Sbjct 157 AFDYSGYGFSSGTQSEKNMYADVRAVYEHI---LKTRPDKKIVVIGYSIGTTAAVDLAAS 213
Query 151 --EEVGGLILQSALLSVHRVALRLRLTFPGDLFVNIKKIKKVKCPVFCIHGANDEVVPIH 208
+ + G++L + L S ALR+ P I KI + V HG +D+ +P+
Sbjct 214 NPDRLVGVVLIAPLTS----ALRMFCNNPDKETTCIDKICHINTRVLICHGDHDQRIPMT 269
Query 209 HGIELYKRARVRVSPLWVGGAGHNNVEIVAGR--EFLLAISRFLR 251
HG+ LY+ + V PL V GA H++ I++G E I+ F+R
Sbjct 270 HGMALYENLKNPVPPLIVHGANHHS--IISGEYIEVFTRIASFMR 312
> CE27315
Length=490
Score = 80.5 bits (197), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 48/157 (30%), Positives = 79/157 (50%), Gaps = 9/157 (5%)
Query 62 LLYSHGNAEDI---FLSFSFFLHLSRVCRADVLLYEYVGYGCSSGKPSERGVYESVEAAY 118
+++S N+ D+ + F ++ + D+L+Y+Y GYG S G +E+ VY +VEA
Sbjct 242 IIFSQPNSSDLGCCLMMDPNFADIADFLQCDLLIYDYPGYGVSEGTTNEKNVYAAVEAVM 301
Query 119 KYLTEELHIHPSKIVAYGRSLGSGPSVHLCASEEVGGLILQSALLSVHRVALR----LRL 174
KY L KI+ G SLG+ VH+ +V ++L + S R+ R +R
Sbjct 302 KYAMGTLGYSQDKIILIGFSLGTAAMVHVAEMYKVAAVVLIAPFTSFFRIVCRRPSIIRP 361
Query 175 TFPGDLFVNIKKIKKVKCPVFCIHGANDEVVPIHHGI 211
F D+F +++K K + P HG D +V HG+
Sbjct 362 WF--DMFPSLEKSKGIGSPTLICHGEKDYIVGHEHGV 396
> Hs14249592
Length=201
Score = 72.4 bits (176), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 47/174 (27%), Positives = 88/174 (50%), Gaps = 14/174 (8%)
Query 94 EYVGYGCSSGKPSERGVYESVEAAYKYLTEELHIHPSKIVAYGRSLGSGPSVHLCA--SE 151
+Y GYG S G+ SE G+Y EA Y+ + +KI +GRSLG ++HL + S
Sbjct 13 DYRGYGKSEGEASEEGLYLDSEAVLDYVMTRPDLDKTKIFLFGRSLGGAVAIHLASENSH 72
Query 152 EVGGLILQSALLSVHRVALRLRLTFP---------GDLFVNIKKIKKVKCPVFCIHGAND 202
+ +++++ LS+ +A L FP + F++ +KI + + P I G +D
Sbjct 73 RISAIMVENTFLSIPHMASTLFSFFPMRYLPLWCYKNKFLSYRKISQCRMPSLFISGLSD 132
Query 203 EVVPIHHGIELYKRARVRVSPLWVGGAG-HNNVEIVAGREFLLAISRFLRLLLR 255
+++P +LY+ + R L + G HN+ G + A+ +F++ +++
Sbjct 133 QLIPPVMMKQLYELSPSRTKRLAIFPDGTHNDTWQCQG--YFTALEQFIKEVVK 184
> At5g20520
Length=340
Score = 70.5 bits (171), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 46/152 (30%), Positives = 71/152 (46%), Gaps = 20/152 (13%)
Query 89 DVLLYEYVGYGCSSGKPSERGVYESVEAAYKYLTEELHIHPSKIVAYGRSLGSGPSVHLC 148
+V + Y GYG S G PS++G+ + +AA +L+ I S+IV +GRSLG L
Sbjct 137 NVFMLSYRGYGASEGYPSQQGIIKDAQAALDHLSGRTDIDTSRIVVFGRSLGGAVGAVLT 196
Query 149 AS--EEVGGLILQSALLSVHRVA------------------LRLRLTFPGDLFVNIKKIK 188
+ ++V LIL++ S+ +A L+L + I I
Sbjct 197 KNNPDKVSALILENTFTSILDMAGVLLPFLKWFIGGSGTKSLKLLNFVVRSPWKTIDAIA 256
Query 189 KVKCPVFCIHGANDEVVPIHHGIELYKRARVR 220
++K PV + G DE+VP H LY +A R
Sbjct 257 EIKQPVLFLSGLQDEMVPPFHMKMLYAKAAAR 288
> CE26316
Length=345
Score = 70.1 bits (170), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 50/188 (26%), Positives = 87/188 (46%), Gaps = 28/188 (14%)
Query 57 SGECTLLYSHGNAED-IFLSFSFFLHLSRVCRADVLLYEYVGYGCSSGKPSERGVYESVE 115
S + Y+HGN+ D F +L C V+ ++Y GYG S G P+E+G+ E +
Sbjct 111 SENQIIFYAHGNSFDRTFYHRVEMYNLLSDCNYHVVCFDYRGYGDSEGTPTEKGIVEDTK 170
Query 116 AAYKYLTEELHIHPSKIVAYGRSLGSGPS---VHLCASEEVG--GLILQSALLSVHRVAL 170
Y++L E P ++ +G S+G+G S V + E+ GLIL+S ++
Sbjct 171 TVYEWLKENCGKTP--VIVWGHSMGTGVSCKLVQDLSREQQPPCGLILESPFNNLKDAVT 228
Query 171 R----LRLTFPGDLFVN----------------IKKIKKVKCPVFCIHGANDEVVPIHHG 210
++ D V+ K+I+ V CP+ +H +D+++P+ G
Sbjct 229 NHPIFTVFSWMNDFMVDHIIIRPLNSVGLTMRSDKRIRLVSCPIIILHAEDDKILPVKLG 288
Query 211 IELYKRAR 218
LY+ A+
Sbjct 289 RALYEAAK 296
> 7295869
Length=338
Score = 69.3 bits (168), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 64/203 (31%), Positives = 90/203 (44%), Gaps = 15/203 (7%)
Query 61 TLLYSHGNAEDIFLSFSFFLHLSRVCRADVLLYEYVGYGCSSGKPSERGVYESVEAAYKY 120
TLLY HGNA ++ + +VL+ EY GYG S+G P+ERG+ AA Y
Sbjct 110 TLLYFHGNAGNMGHRMQNVWGIYHHLHCNVLMVEYRGYGLSTGVPTERGLVTDARAAIDY 169
Query 121 LTEELHIHPSKIVAYGRSLGSGPSVHLCASEEVGG----LILQSALLSVHRVALRLRLT- 175
L + S+++ +GRSLG V + A G I+++ S+ +A+ L
Sbjct 170 LHTRHDLDHSQLILFGRSLGGAVVVDVAADTVYGQKLMCAIVENTFSSIPEMAVELVHPA 229
Query 176 ---FPGDLFVN----IKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLW-VG 227
P LF N + KI K P I G D +VP LY + + L
Sbjct 230 VKYIPNLLFKNKYHSMSKIGKCSVPFLFISGLADNLVPPRMMRALYTKCGSEIKRLLEFP 289
Query 228 GAGHNNVEIVAGREFLLAISRFL 250
G HN+ IV G + AI FL
Sbjct 290 GGSHNDTWIVDG--YYQAIGGFL 310
> SPBC32H8.03
Length=299
Score = 68.2 bits (165), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 48/167 (28%), Positives = 79/167 (47%), Gaps = 16/167 (9%)
Query 61 TLLYSHGNAEDIFLSFSFFLHLSRVCRADVLLYEYVGYGCSSGKPSERGVYESVEAAYKY 120
TLLY H NA ++ +V + Y GYG S+G PSE G+ + A +Y
Sbjct 91 TLLYFHANAGNMGHRLPIARVFYSALNMNVFIISYRGYGKSTGSPSEAGLKIDSQTALEY 150
Query 121 LTEELHIHPSKIVAYGRSLGSGPSVHLCASEE--VGGLILQSALLSVHRVALRLRLTFP- 177
L E +KIV YG+S+G ++ L A + + LIL++ S+ + + FP
Sbjct 151 LMEHPICSKTKIVVYGQSIGGAVAIALTAKNQDRISALILENTFTSIKDM---IPTVFPY 207
Query 178 ---------GDLFVNIKKIKKV-KCPVFCIHGANDEVVPIHHGIELY 214
+++ + +I+K+ K PV + G DE+VP + L+
Sbjct 208 GGSIISRFCTEIWSSQDEIRKIKKLPVLFLSGEKDEIVPPPQMVLLF 254
> ECU09g0730
Length=326
Score = 62.0 bits (149), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 47/188 (25%), Positives = 81/188 (43%), Gaps = 10/188 (5%)
Query 51 LLFVGRSGECTLLYSHGNAEDIFLSFSFFLHLSRVCRADVLLYEYVGYGCSSGKPSERGV 110
L + + ++Y GN F +LS+ R + + Y G + PSERG+
Sbjct 117 LYLIDQMSSTDIIYLPGNFVVTREHIEFCKYLSKALRCNTVSMIYRGIAGNPHVPSERGI 176
Query 111 YESVEAAYKYLTEELHIHPSKIVAYGRSLGSGPSVHLCASEEVGGLILQSALLSVHRVAL 170
E V A ++L+ ++ V G S+G+ + L V L+L + +S+ V
Sbjct 177 VEDVSTASEWLSR----RSTRKVVVGFSIGAAVGIRLAGMCRVDALVLVNPFISLREVVS 232
Query 171 RLRLT-----FPGDLFVNIKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLW 225
+ L D + N+ +K + PV+ + ++DE+VP H L KR R +
Sbjct 233 SIPLGRVLKHLVVDEWSNMNGVKDIDVPVYFVVSSDDEIVPESHADALIKRTR-HPRKIV 291
Query 226 VGGAGHNN 233
+ A HN
Sbjct 292 IRNADHNE 299
> YNL320w
Length=284
Score = 58.9 bits (141), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 49/175 (28%), Positives = 80/175 (45%), Gaps = 24/175 (13%)
Query 90 VLLYEYVGYGCSSGKPSERGVYESVEAAYKYLTEELHIHPSKIVAYGRSLGSGPSVHLCA 149
V +Y Y GYG S G PSE+G+ + +L+ + K+V YGRSLG ++++ +
Sbjct 109 VFIYSYRGYGNSEGSPSEKGLKLDADCVISHLSTDSFHSKRKLVLYGRSLGGANALYIAS 168
Query 150 S--EEVGGLILQSALLSVHRV-----ALRLRLTFPGDLFVNIKKIK---KVKCPVFCIHG 199
+ G+IL++ LS+ +V L R T N + + + P + G
Sbjct 169 KFRDLCDGVILENTFLSIRKVIPYIFPLLKRFTLLCHEIWNSEGLMGSCSSETPFLFLSG 228
Query 200 ANDEVVPIHHGIELYK-----RARVRVSPLWVGGAGHNNVEIVAG-----REFLL 244
DE+VP H +LY+ ++ PL HN+ I G R+FL+
Sbjct 229 LKDEIVPPFHMRKLYETCPSSNKKIFEFPL----GSHNDTIIQDGYWDIIRDFLI 279
> Hs22056657
Length=124
Score = 57.4 bits (137), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 40/132 (30%), Positives = 61/132 (46%), Gaps = 24/132 (18%)
Query 135 YGRSLGSGPSVHLCASEEVGGLILQSALLSVHRVAL----RLRLTFPGDLFV-------N 183
Y +S+G+ P+V L + E ++L S L VA R + P F
Sbjct 2 YRQSIGTVPTVDLASRYESAAVVLHSPLTLDLSVAFPTPRRPTASTPSPKFSPAPRPSRR 61
Query 184 IKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNVEIVAGREFL 243
I+K+ K+ PV IHG DE +R V PLWV GAGH ++++ + ++L
Sbjct 62 IQKVSKITSPVLIIHGTKDE-----------ERCPKAVEPLWVEGAGHKDIQLYS--QYL 108
Query 244 LAISRFLRLLLR 255
+ RF+ LR
Sbjct 109 ERLRRFISQELR 120
> Hs20559602
Length=404
Score = 57.4 bits (137), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 42/148 (28%), Positives = 67/148 (45%), Gaps = 25/148 (16%)
Query 90 VLLYEYVGYGCSSGKPSERGVYESVEAAYKYLTEELHIHPSKIVAYGRSLGSGPSVHL-- 147
V+ ++Y G+G S G PSERG+ + ++ +P + +G SLG+G + +L
Sbjct 200 VVTFDYRGWGDSVGTPSERGMTYDALHVFDWIKARSGDNP--VYIWGHSLGTGVATNLVR 257
Query 148 --CASE-EVGGLILQSALLSVHRVALRLRLT-----FPG-DLF------------VNIKK 186
C E LIL+S ++ A + FPG D F N +
Sbjct 258 RLCERETPPDALILESPFTNIREEAKSHPFSVIYRYFPGFDWFFLDPITSSGIKFANDEN 317
Query 187 IKKVKCPVFCIHGANDEVVPIHHGIELY 214
+K + CP+ +H +D VVP G +LY
Sbjct 318 VKHISCPLLILHAEDDPVVPFQLGRKLY 345
> Hs18426888
Length=166
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 25/61 (40%), Positives = 40/61 (65%), Gaps = 1/61 (1%)
Query 61 TLLYSHGNAEDIFLSFSFFLHLSRVCRADVLLYEYVGYGCSSGKPSERGVYESVEAAYKY 120
T+ +SHGNA D+ SF++ L ++ Y+Y GYG S+G+PSER +Y ++AA++
Sbjct 104 TVFFSHGNAVDLSQMSSFYIGLGSRLHCNIF-YDYSGYGASAGRPSERNLYADIDAAWQA 162
Query 121 L 121
L
Sbjct 163 L 163
> Hs22065370
Length=93
Score = 40.4 bits (93), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 23/62 (37%), Positives = 35/62 (56%), Gaps = 1/62 (1%)
Query 127 IHPSKIVAYGRSLGSGPSVHLCASEEVGGLILQSALLSVHRVAL-RLRLTFPGDLFVNIK 185
I P I+ Y +S+G+ P+V L + E ++L S L S RVA + T+ D F NI+
Sbjct 31 ISPDSIILYRQSIGTVPTVDLASRYECAAVVLHSPLTSGMRVAFPDTKTTYCFDAFPNIE 90
Query 186 KI 187
K+
Sbjct 91 KV 92
> Hs22048162
Length=90
Score = 36.6 bits (83), Expect = 0.058, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 25/39 (64%), Gaps = 0/39 (0%)
Query 180 LFVNIKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRAR 218
+F N + +K + P+ +HG +D VP+ +G +LY+ AR
Sbjct 10 IFPNDENVKFLSSPLLILHGEDDRTVPLEYGKKLYEIAR 48
> Hs22061602
Length=1246
Score = 36.2 bits (82), Expect = 0.059, Method: Compositional matrix adjust.
Identities = 22/84 (26%), Positives = 43/84 (51%), Gaps = 1/84 (1%)
Query 90 VLLYEYVGYGCSSGKPSERGVYESVEAAYKYLTEELHIHPSKIVAYGRSLGSGPSVHLCA 149
VL + + G+G S+G P + +++ +Y LH P+ +V YG S+G +
Sbjct 976 VLGWNHPGFGSSTGVPFPQHDANAMDVVVEYALHRLHFPPAHLVVYGWSVGGFTATWATM 1035
Query 150 S-EEVGGLILQSALLSVHRVALRL 172
+ E+G L+L + + +AL++
Sbjct 1036 TYPELGALVLDATFDDLVPLALKV 1059
> CE01231
Length=629
Score = 35.8 bits (81), Expect = 0.087, Method: Compositional matrix adjust.
Identities = 16/48 (33%), Positives = 26/48 (54%), Gaps = 3/48 (6%)
Query 187 IKKVKCPVFCIHGANDEVVPIHHGIELYKRAR---VRVSPLWVGGAGH 231
I K++ P+ +HG D VVP+ I +Y++ R V + + G GH
Sbjct 542 IDKIRTPIAFLHGREDTVVPMSQSITMYEKIRASGVTTALMLFDGEGH 589
> YLR118c
Length=227
Score = 34.7 bits (78), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 33/124 (26%), Positives = 56/124 (45%), Gaps = 16/124 (12%)
Query 93 YEYVGYGCSSGKPSERGVYESVEAAYKYLTEELH--IHPSKIVAYGRSLGSGPSVHLCAS 150
++ + + S K G S+ + K + +E+ I P +I+ G S G+ ++ +
Sbjct 72 FDILEWDPSFSKVDSDGFMNSLNSIEKTVKQEIDKGIKPEQIIIGGFSQGAALALATSVT 131
Query 151 --EEVGGLILQSALLSVHRVALRLRLTFPGDLFVNIKKIKKVKCPVFCIHGANDEVVPIH 208
++GG++ S S+ PG L + I VK P+F HG D VVPI
Sbjct 132 LPWKIGGIVALSGFCSI-----------PGILKQHKNGIN-VKTPIFHGHGDMDPVVPIG 179
Query 209 HGIE 212
GI+
Sbjct 180 LGIK 183
> CE14317
Length=445
Score = 34.3 bits (77), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 19/65 (29%), Positives = 37/65 (56%), Gaps = 4/65 (6%)
Query 165 VHRVALRLRLTFPGDLFVNIKKIKKVKCPV---FCIH-GANDEVVPIHHGIELYKRARVR 220
++ +A+R+ + + VN +++K+ V C H G +E++P HH +E+ + A VR
Sbjct 319 INVMAMRVLMKMAAEKSVNEQQLKEALETVIGGLCKHTGRKNELIPDHHPLEISRNAAVR 378
Query 221 VSPLW 225
PL+
Sbjct 379 QYPLF 383
> CE08878
Length=154
Score = 34.3 bits (77), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 11/31 (35%), Positives = 20/31 (64%), Gaps = 0/31 (0%)
Query 186 KIKKVKCPVFCIHGANDEVVPIHHGIELYKR 216
K+ ++CPV +HG D +VP+ + +L +R
Sbjct 77 KVIDIQCPVLTVHGIEDNIVPVENSKKLMQR 107
> Hs13640732
Length=732
Score = 34.3 bits (77), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 14/41 (34%), Positives = 22/41 (53%), Gaps = 0/41 (0%)
Query 184 IKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPL 224
I+ I +VK P+ + G D VP G+E Y+ + R P+
Sbjct 657 IRYIPQVKTPLLLMLGQEDRRVPFKQGMEYYRALKTRNVPV 697
> Hs9951917
Length=732
Score = 34.3 bits (77), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 14/41 (34%), Positives = 22/41 (53%), Gaps = 0/41 (0%)
Query 184 IKKIKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPL 224
I+ I +VK P+ + G D VP G+E Y+ + R P+
Sbjct 657 IRYIPQVKTPLLLMLGQEDRRVPFKQGMEYYRALKTRNVPV 697
> CE16858
Length=561
Score = 33.9 bits (76), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 19/65 (29%), Positives = 37/65 (56%), Gaps = 4/65 (6%)
Query 165 VHRVALRLRLTFPGDLFVNIKKIKKVKCPV---FCIH-GANDEVVPIHHGIELYKRARVR 220
++ +A+R+ + + VN +++K+ V C H G +E++P HH +E+ + A VR
Sbjct 435 INVMAMRVLMKMAAEKSVNEQQLKEALETVIGGLCKHTGRKNELIPDHHPLEISRNAAVR 494
Query 221 VSPLW 225
PL+
Sbjct 495 QYPLF 499
> CE27390
Length=223
Score = 33.9 bits (76), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 28/114 (24%), Positives = 52/114 (45%), Gaps = 19/114 (16%)
Query 102 SGKPSERGVYESVEAAYKYLTEEL--HIHPSKIVAYGRSLGSGPSVH--LCASEEVGGLI 157
+ + E+G+ + + ++ + E+ I S+I G S+G +++ L +++GG++
Sbjct 80 NAQEDEQGINRATQYVHQLIDAEVAAGIPASRIAVGGFSMGGALAIYAGLTYPQKLGGIV 139
Query 158 -LQSALLSVHRVALRLRLTFPGDLFVNIKKIKKVKCPVFCIHGANDEVVPIHHG 210
L S L R FPG N P+F HG +D +VP+ G
Sbjct 140 GLSSFFLQ--------RTKFPGSFTAN------NATPIFLGHGTDDFLVPLQFG 179
> Hs15100151
Length=558
Score = 33.1 bits (74), Expect = 0.60, Method: Compositional matrix adjust.
Identities = 25/127 (19%), Positives = 53/127 (41%), Gaps = 11/127 (8%)
Query 90 VLLYEYVGYGCSSGKPSERGVYESVEAAYKYLTEELHIHPSKIVAYGRSLGSGPSVHLCA 149
VL + + G+ S+G P + +++ ++ L P I+ Y S+G +
Sbjct 307 VLGWNHPGFAGSTGVPFPQNEANAMDVVVQFAIHRLGFQPQDIIIYAWSIGGFTATWAAM 366
Query 150 S-EEVGGLILQSALLSVHRVALRLRLTFPGDLFV----------NIKKIKKVKCPVFCIH 198
S +V +IL ++ + +AL++ L N +++ + + PV I
Sbjct 367 SYPDVSAMILDASFDDLVPLALKVMPDSWRGLVTRTVRQHLNLNNAEQLCRYQGPVLLIR 426
Query 199 GANDEVV 205
DE++
Sbjct 427 RTKDEII 433
> At1g64670
Length=469
Score = 32.3 bits (72), Expect = 0.87, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 35/71 (49%), Gaps = 11/71 (15%)
Query 188 KKVKCPVFCIHGANDEVVPIH--HGIE-LYKRARVRVSPLWVGGAGHNNVEIVAGREFLL 244
V C V HG DE++P+ +G++ RAR+ V P +++ IV GR+
Sbjct 396 DNVDCEVAVFHGGRDELIPVECSYGVKRKVPRARIHVVP------DKDHITIVVGRQKEF 449
Query 245 AISRFLRLLLR 255
A R L L+ R
Sbjct 450 A--RELELIWR 458
> 7294700_2
Length=211
Score = 32.0 bits (71), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 28/112 (25%), Positives = 49/112 (43%), Gaps = 17/112 (15%)
Query 103 GKPSERGVYESVEAAYKYLTEELH--IHPSKIVAYGRSLGSGPSVH--LCASEEVGGLIL 158
G E G+ + ++ + + +E+ I ++IV G S G +++ L + + G++
Sbjct 49 GPEDEPGIQSARDSVHGMIQKEISAGIPANRIVLGGFSQGGALALYSALTYDQPLAGVVA 108
Query 159 QSALLSVHRVALRLRLTFPGDLFVNIKKIKKVKCPVFCIHGANDEVVPIHHG 210
S L +H+ FPG K+ P+F HG D VVP G
Sbjct 109 LSCWLPLHK-------QFPG------AKVNSDDVPIFQAHGDYDPVVPYKFG 147
> 7292468
Length=524
Score = 32.0 bits (71), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 31/135 (22%), Positives = 57/135 (42%), Gaps = 13/135 (9%)
Query 90 VLLYEYVGYGCSSGKPSERGVYESVEAAYKYLTEELHIHPSKIVAYGRSLGSGPSVHLCA 149
VL + + G+ S+G P +++A ++ L I+ YG S+G G S A
Sbjct 271 VLGWNHPGFAGSTGTPHPHQDKNAIDAVVQFAINNLRFPVEDIILYGWSIG-GFSTLYAA 329
Query 150 S--EEVGGLILQSALLSVHRVAL-RLRLTFPGDLFVNIKK---------IKKVKCPVFCI 197
S +V G++L + V +A+ R+ G + V I+ + P+ I
Sbjct 330 SVYPDVKGVVLDATFDDVLYLAVPRMPAALAGIVKVAIRNYCNLNNAELANEFNGPISFI 389
Query 198 HGANDEVVPIHHGIE 212
DE++ + I+
Sbjct 390 RRTEDEIIAEDNHID 404
> Hs9966764
Length=231
Score = 31.6 bits (70), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 40/161 (24%), Positives = 67/161 (41%), Gaps = 17/161 (10%)
Query 99 GCSSGKP-SERGVYESVEAAYKYLTEELH--IHPSKIVAYGRSLGSGPSVH--LCASEEV 153
G S P E G+ ++ E + E+ I ++IV G S G S++ L +
Sbjct 80 GLSPDAPEDEAGIKKAAENIKALIEHEMKNGIPANRIVLGGFSQGGALSLYTALTCPHPL 139
Query 154 GGLILQSALLSVHRVALRLRLTFPGDLFVNIKKIKKVKCPVFCIHGANDEVVPIHHGIEL 213
G++ S L +HR FP + K + ++C HG D +VP+ G
Sbjct 140 AGIVALSCWLPLHRA-------FPQAANGSAKDLAILQC-----HGELDPMVPVRFGALT 187
Query 214 YKRARVRVSPLWVGGAGHNNVEIVAGREFLLAISRFLRLLL 254
++ R V+P V + V + + + A+ FL LL
Sbjct 188 AEKLRSVVTPARVQFKTYPGVMHSSCPQEMAAVKEFLEKLL 228
> At4g24140
Length=498
Score = 31.6 bits (70), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 20/70 (28%), Positives = 38/70 (54%), Gaps = 10/70 (14%)
Query 179 DLFVNIKKIKKVKCPVFCIHGANDEVVPI---HHGIELYKRARVRVSPLWVGGAGHNNVE 235
D +++I + K+KC V HG +DE++P+ ++ + RARV+V +++
Sbjct 424 DTYLDIVR-DKLKCNVTIFHGGDDELIPVECSYNVKQRIPRARVKVI------EHKDHIT 476
Query 236 IVAGREFLLA 245
+V GR+ A
Sbjct 477 MVVGRQDEFA 486
> Hs4757862
Length=274
Score = 31.6 bits (70), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 19/65 (29%), Positives = 32/65 (49%), Gaps = 2/65 (3%)
Query 187 IKKVKCPVFCIHGANDEVVPIHHGIELYKRARVRVSPLWVGGAGHNNVEIVAGREFLLAI 246
+ +V+CP +HG D +VP H ++K V+ S L + G +N+ + EF
Sbjct 212 LPRVQCPALIVHGEKDPLVPRFHADFIHK--HVKGSRLHLMPEGKHNLHLRFADEFNKLA 269
Query 247 SRFLR 251
FL+
Sbjct 270 EDFLQ 274
Lambda K H
0.329 0.143 0.465
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5294113934
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40