bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0608_orf1
Length=171
Score E
Sequences producing significant alignments: (Bits) Value
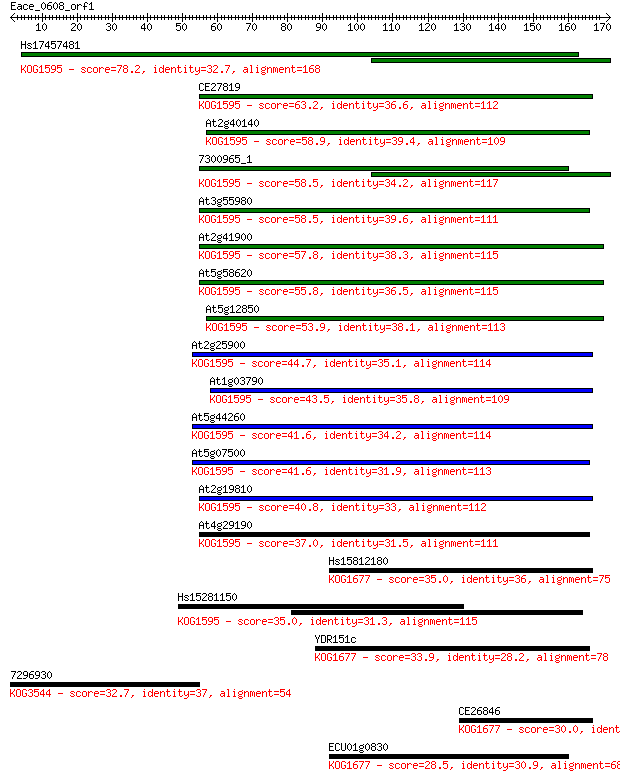
Hs17457481 78.2 8e-15
CE27819 63.2 3e-10
At2g40140 58.9 4e-09
7300965_1 58.5 6e-09
At3g55980 58.5 6e-09
At2g41900 57.8 1e-08
At5g58620 55.8 4e-08
At5g12850 53.9 2e-07
At2g25900 44.7 1e-04
At1g03790 43.5 2e-04
At5g44260 41.6 7e-04
At5g07500 41.6 9e-04
At2g19810 40.8 0.001
At4g29190 37.0 0.019
Hs15812180 35.0 0.078
Hs15281150 35.0 0.082
YDR151c 33.9 0.15
7296930 32.7 0.35
CE26846 30.0 2.1
ECU01g0830 28.5 6.2
> Hs17457481
Length=412
Score = 78.2 bits (191), Expect = 8e-15, Method: Compositional matrix adjust.
Identities = 53/163 (32%), Positives = 77/163 (47%), Gaps = 13/163 (7%)
Query 4 AAAEISAAAGSSSGASSGASAGAAAGAAAAAAAAKSTRLSWSCGDTEGSLNLSSFKVFPC 63
A + + G+ G SAGAA+ A ++ R W E + L ++K PC
Sbjct 168 AMEALQNGQTTVEGSIEGQSAGAASHAMIEKILSEEPR--WQ----ETAYVLGNYKTEPC 221
Query 64 HQKHSAAHDKKYCPFYHNFRDKRRFPV--TYRAEQCE--EHFDSDSASLQCSKGDACEKS 119
+ CP+YHN +D+RR P YR+ C +H D +C GDAC+
Sbjct 222 KKPPRLCRQGYACPYYHNSKDRRRSPRKHKYRSSPCPNVKHGDEWGDPGKCENGDACQYC 281
Query 120 HNRHELLYHPTIFKQRFCSSYATRNGTERCGRGNFCAFAHSRE 162
H R E +HP I+K C+ ++G+ C RG FCAFAH +
Sbjct 282 HTRTEQQFHPEIYKSTKCND-MQQSGS--CPRGPFCAFAHVEQ 321
Score = 32.3 bits (72), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 22/72 (30%), Positives = 32/72 (44%), Gaps = 6/72 (8%)
Query 104 DSASLQCSKGDACEKSHNRH---ELLYHPTIFKQRFCSSYATRNGTERCGR-GNFCAFAH 159
D A+ C +GD C H E YH +K C G C + G CAFAH
Sbjct 94 DEATGLCPEGDECPFLHRTTGDTERRYHLRYYKTGICIHETDSKGN--CTKNGLHCAFAH 151
Query 160 SREEVRAPLFSV 171
++R+P++ +
Sbjct 152 GPHDLRSPVYDI 163
> CE27819
Length=704
Score = 63.2 bits (152), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 41/120 (34%), Positives = 53/120 (44%), Gaps = 15/120 (12%)
Query 55 LSSFKVFPCHQKHSAAHDKKYCPFYHNFRDKRRFPVTYR--------AEQCEEHFDSDSA 106
LS +K C + CPFYHN +D+RR P Y+ A+ +E D D
Sbjct 187 LSCYKTEQCRKPARLCRQGYACPFYHNSKDRRRPPALYKYRSTPCPAAKTIDEWLDPDI- 245
Query 107 SLQCSKGDACEKSHNRHELLYHPTIFKQRFCSSYATRNGTERCGRGNFCAFAHSREEVRA 166
C GD C+ H R E +HP I+K C+ C R FCAFAH E+ A
Sbjct 246 ---CEAGDNCQYCHTRTEQQFHPEIYKSTKCNDMLEHG---YCPRAVFCAFAHHDSELHA 299
> At2g40140
Length=597
Score = 58.9 bits (141), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 43/114 (37%), Positives = 53/114 (46%), Gaps = 21/114 (18%)
Query 57 SFKVFPCHQKHSAAHDKKYCPFYH-----NFRDKRRFPVTYRAEQCEEHFDSDSASLQCS 111
SFKV PC + +S HD CPF H RD R++P Y C E F S C
Sbjct 219 SFKVKPCSRAYS--HDWTECPFVHPGENARRRDPRKYP--YTCVPCPE-FRKGS----CP 269
Query 112 KGDACEKSHNRHELLYHPTIFKQRFCSSYATRNGTERCGRGNFCAFAHSREEVR 165
KGD+CE +H E HP ++ R C C R C FAH R+E+R
Sbjct 270 KGDSCEYAHGVFESWLHPAQYRTRLCKDETG------CAR-RVCFFAHRRDELR 316
> 7300965_1
Length=541
Score = 58.5 bits (140), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 38/109 (34%), Positives = 50/109 (45%), Gaps = 7/109 (6%)
Query 55 LSSFKVFPCHQKHSAAHDKKYCPFYHNFRDKRRFP--VTYRAEQCE--EHFDSDSASLQC 110
L+++K PC + CP YHN +DKRR P YR+ C +H + C
Sbjct 207 LANYKTEPCKRPPRLCRQGYACPQYHNSKDKRRSPRKYKYRSTPCPNVKHGEEWGEPGNC 266
Query 111 SKGDACEKSHNRHELLYHPTIFKQRFCSSYATRNGTERCGRGNFCAFAH 159
GD C+ H R E +HP I+K C+ C R FCAFAH
Sbjct 267 EAGDNCQYCHTRTEQQFHPEIYKSTKCNDVQQ---AGYCPRSVFCAFAH 312
Score = 29.3 bits (64), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 21/77 (27%), Positives = 30/77 (38%), Gaps = 16/77 (20%)
Query 104 DSASLQCSKGDACEKSHNR---HELLYHPTIFKQRFCS------SYATRNGTERCGRGNF 154
D + C +GD C H E YH +K C Y +NG
Sbjct 96 DETTGICPEGDECPYLHRTAGDTERRYHLRYYKTCMCVHDTDSRGYCVKNGLH------- 148
Query 155 CAFAHSREEVRAPLFSV 171
CAFAH ++ R P++ +
Sbjct 149 CAFAHGMQDQRPPVYDI 165
> At3g55980
Length=586
Score = 58.5 bits (140), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 44/116 (37%), Positives = 53/116 (45%), Gaps = 21/116 (18%)
Query 55 LSSFKVFPCHQKHSAAHDKKYCPFYH-----NFRDKRRFPVTYRAEQCEEHFDSDSASLQ 109
+ SFKV PC + +S HD C F H RD R++P Y C E F S
Sbjct 220 MYSFKVKPCSRAYS--HDWTECAFVHPGENARRRDPRKYP--YTCVPCPE-FRKGS---- 270
Query 110 CSKGDACEKSHNRHELLYHPTIFKQRFCSSYATRNGTERCGRGNFCAFAHSREEVR 165
C KGD+CE +H E HP +K R C C R C FAH REE+R
Sbjct 271 CPKGDSCEYAHGVFESWLHPAQYKTRLCKDETG------CAR-KVCFFAHKREEMR 319
> At2g41900
Length=727
Score = 57.8 bits (138), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 44/120 (36%), Positives = 57/120 (47%), Gaps = 22/120 (18%)
Query 55 LSSFKVFPCHQKHSAAHDKKYCPFYH-----NFRDKRRFPVTYRAEQCEEHFDSDSASLQ 109
+ SFKV PC + +S HD CPF H RD R+F Y C D
Sbjct 275 MYSFKVRPCSRAYS--HDWTECPFVHPGENARRRDPRKF--HYSCVPCP-----DFRKGA 325
Query 110 CSKGDACEKSHNRHELLYHPTIFKQRFCSSYATRNGTERCGRGNFCAFAHSREEVRAPLF 169
C +GD CE +H E HP ++ R C ++GT C R C FAH+ EE+R PL+
Sbjct 326 CRRGDMCEYAHGVFECWLHPAQYRTRLC-----KDGTG-CAR-RVCFFAHTPEELR-PLY 377
> At5g58620
Length=607
Score = 55.8 bits (133), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 42/120 (35%), Positives = 55/120 (45%), Gaps = 22/120 (18%)
Query 55 LSSFKVFPCHQKHSAAHDKKYCPFYH-----NFRDKRRFPVTYRAEQCEEHFDSDSASLQ 109
+ +FK+ PC + +S HD CPF H RD R++ Y C E F S
Sbjct 212 MYAFKIKPCSRAYS--HDWTECPFVHPGENARRRDPRKY--HYSCVPCPE-FRKGS---- 262
Query 110 CSKGDACEKSHNRHELLYHPTIFKQRFCSSYATRNGTERCGRGNFCAFAHSREEVRAPLF 169
CS+GD CE +H E HP ++ R C C R C FAH EE+R PL+
Sbjct 263 CSRGDTCEYAHGIFECWLHPAQYRTRLCKDETN------CSR-RVCFFAHKPEELR-PLY 314
> At5g12850
Length=706
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 43/118 (36%), Positives = 57/118 (48%), Gaps = 22/118 (18%)
Query 57 SFKVFPCHQKHSAAHDKKYCPFYH-----NFRDKRRFPVTYRAEQCEEHFDSDSASLQCS 111
SFK+ PC + +S HD CPF H RD R+F Y C + F S C
Sbjct 262 SFKIRPCSRAYS--HDWTECPFAHPGENARRRDPRKF--HYTCVPCPD-FKKGS----CK 312
Query 112 KGDACEKSHNRHELLYHPTIFKQRFCSSYATRNGTERCGRGNFCAFAHSREEVRAPLF 169
+GD CE +H E HP ++ R C ++G C R C FAH+ EE+R PL+
Sbjct 313 QGDMCEYAHGVFECWLHPAQYRTRLC-----KDGMG-CNR-RVCFFAHANEELR-PLY 362
> At2g25900
Length=315
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 40/119 (33%), Positives = 55/119 (46%), Gaps = 21/119 (17%)
Query 53 LNLSSFKVFPCHQKHSAAHDKKYCPFYH-----NFRDKRRFPVTYRAEQCEEHFDSDSAS 107
+ FK+ C + S HD CPF H RD R+F Y C E F S
Sbjct 91 FRIYEFKIRRCARGRS--HDWTECPFAHPGEKARRRDPRKF--HYSGTACPE-FRKGS-- 143
Query 108 LQCSKGDACEKSHNRHELLYHPTIFKQRFCSSYATRNGTERCGRGNFCAFAHSREEVRA 166
C +GD+CE SH E HP+ ++ + C ++GT C R C FAH+ E++R
Sbjct 144 --CRRGDSCEFSHGVFECWLHPSRYRTQPC-----KDGTS-C-RRRICFFAHTTEQLRV 193
> At1g03790
Length=393
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 39/114 (34%), Positives = 51/114 (44%), Gaps = 20/114 (17%)
Query 58 FKVFPCHQKHSAAHDKKYCPFYH-----NFRDKRRFPVTYRAEQCEEHFDSDSASLQCSK 112
FK+ C + S +HD CPF H RD RRF Y E C E CS+
Sbjct 88 FKIRRCTR--SRSHDWTDCPFAHPGEKARRRDPRRF--QYSGEVCPEF----RRGGDCSR 139
Query 113 GDACEKSHNRHELLYHPTIFKQRFCSSYATRNGTERCGRGNFCAFAHSREEVRA 166
GD CE +H E HP ++ C ++G + C R C FAHS ++R
Sbjct 140 GDDCEFAHGVFECWLHPIRYRTEAC-----KDG-KHCKR-KVCFFAHSPRQLRV 186
> At5g44260
Length=381
Score = 41.6 bits (96), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 39/119 (32%), Positives = 53/119 (44%), Gaps = 20/119 (16%)
Query 53 LNLSSFKVFPCHQKHSAAHDKKYCPFYH-----NFRDKRRFPVTYRAEQCEEHFDSDSAS 107
+ FK+ C + S +HD CPF H RD RRF Y E C E S
Sbjct 61 FRMYEFKIRRCTR--SRSHDWTDCPFSHPGEKARRRDPRRF--HYTGEVCPEF----SRH 112
Query 108 LQCSKGDACEKSHNRHELLYHPTIFKQRFCSSYATRNGTERCGRGNFCAFAHSREEVRA 166
CS+GD C +H E HP+ ++ C ++G + C R C FAHS ++R
Sbjct 113 GDCSRGDECGFAHGVFECWLHPSRYRTEAC-----KDG-KHCKR-KVCFFAHSPRQLRV 164
> At5g07500
Length=245
Score = 41.6 bits (96), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 36/118 (30%), Positives = 52/118 (44%), Gaps = 21/118 (17%)
Query 53 LNLSSFKVFPCHQKHSAAHDKKYCPFYH-----NFRDKRRFPVTYRAEQCEEHFDSDSAS 107
+ ++K+ C + S HD CP+ H RD RR+ TY A C +
Sbjct 53 FRMYAYKIKRCPRTRS--HDWTECPYAHRGEKATRRDPRRY--TYCAVACPAFRNG---- 104
Query 108 LQCSKGDACEKSHNRHELLYHPTIFKQRFCSSYATRNGTERCGRGNFCAFAHSREEVR 165
C +GD+CE +H E HP ++ R C N C R C FAH+ E++R
Sbjct 105 -ACHRGDSCEFAHGVFEYWLHPARYRTRAC------NAGNLCQR-KVCFFAHAPEQLR 154
> At2g19810
Length=359
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 37/117 (31%), Positives = 48/117 (41%), Gaps = 21/117 (17%)
Query 55 LSSFKVFPCHQKHSAAHDKKYCPFYH-----NFRDKRRFPVTYRAEQCEEHFDSDSASLQ 109
+ FKV C + S HD CP+ H RD R+F Y C E
Sbjct 81 MYEFKVRRCARGRS--HDWTECPYAHPGEKARRRDPRKF--HYSGTACPEFRKG-----C 131
Query 110 CSKGDACEKSHNRHELLYHPTIFKQRFCSSYATRNGTERCGRGNFCAFAHSREEVRA 166
C +GDACE SH E HP ++ + C C R C FAHS +++R
Sbjct 132 CKRGDACEFSHGVFECWLHPARYRTQPCKDGGN------C-RRRVCFFAHSPDQIRV 181
> At4g29190
Length=356
Score = 37.0 bits (84), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 35/116 (30%), Positives = 48/116 (41%), Gaps = 21/116 (18%)
Query 55 LSSFKVFPCHQKHSAAHDKKYCPFYH-----NFRDKRRFPVTYRAEQCEEHFDSDSASLQ 109
+ FKV C + S HD CP+ H RD R++ Y C D
Sbjct 82 MYDFKVRRCARGRS--HDWTECPYAHPGEKARRRDPRKY--HYSGTAC-----PDFRKGG 132
Query 110 CSKGDACEKSHNRHELLYHPTIFKQRFCSSYATRNGTERCGRGNFCAFAHSREEVR 165
C KGD+CE +H E HP ++ + C C R C FAHS +++R
Sbjct 133 CKKGDSCEFAHGVFECWLHPARYRTQPCKDGGN------CLR-KICFFAHSPDQLR 181
> Hs15812180
Length=338
Score = 35.0 bits (79), Expect = 0.078, Method: Compositional matrix adjust.
Identities = 27/78 (34%), Positives = 36/78 (46%), Gaps = 11/78 (14%)
Query 92 YRAEQCEEHFDSDSASLQCSKGDACEKSHNRHE---LLYHPTIFKQRFCSSYATRNGTER 148
Y+ E C F+ + A C GD C+ +H HE L HP +K C ++ T
Sbjct 115 YKTELCRP-FEENGA---CKYGDKCQFAHGIHELRSLTRHPK-YKTELCRTFHT---IGF 166
Query 149 CGRGNFCAFAHSREEVRA 166
C G C F H+ EE RA
Sbjct 167 CPYGPRCHFIHNAEERRA 184
> Hs15281150
Length=756
Score = 35.0 bits (79), Expect = 0.082, Method: Compositional matrix adjust.
Identities = 24/85 (28%), Positives = 36/85 (42%), Gaps = 4/85 (4%)
Query 49 TEGSLNLSSFKVFPCHQKHSAAHDKKYCPFYHN--FRDKRRFPVTYRAEQCE--EHFDSD 104
++ + L S+K C + CP YHN R + YR+ C +H D
Sbjct 302 SDANFVLGSYKTEQCPKPPRLCRQGYACPHYHNSRDRRRNPRRFQYRSTPCPSVKHGDEW 361
Query 105 SASLQCSKGDACEKSHNRHELLYHP 129
+C GD C+ H+R E +HP
Sbjct 362 GEPSRCDGGDGCQYCHSRTEQQFHP 386
Score = 32.0 bits (71), Expect = 0.57, Method: Compositional matrix adjust.
Identities = 25/88 (28%), Positives = 38/88 (43%), Gaps = 11/88 (12%)
Query 81 NFRDKRRFPVTYRAEQCEEHFDSDSASLQCSKGDACEKSHNRHELLYHPTIFKQRFCSSY 140
+F D +Y+ EQC + C +G AC HN + +P F+ R
Sbjct 300 HFSDANFVLGSYKTEQCPK------PPRLCRQGYACPHYHNSRDRRRNPRRFQYRSTPCP 353
Query 141 ATRNGTE-----RCGRGNFCAFAHSREE 163
+ ++G E RC G+ C + HSR E
Sbjct 354 SVKHGDEWGEPSRCDGGDGCQYCHSRTE 381
> YDR151c
Length=325
Score = 33.9 bits (76), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 22/85 (25%), Positives = 36/85 (42%), Gaps = 10/85 (11%)
Query 88 FPVTYRAEQCEEHFDSDSASL-------QCSKGDACEKSHNRHELLYHPTIFKQRFCSSY 140
P+T Q DS S L + +K + C ++ + L + T++K C S+
Sbjct 154 LPLTAENLQKLSQVDSQSTGLPYTLPIQKTTKLEPCRRAPLQLPQLVNKTLYKTELCESF 213
Query 141 ATRNGTERCGRGNFCAFAHSREEVR 165
+ C GN C FAH E++
Sbjct 214 TIKG---YCKYGNKCQFAHGLNELK 235
> 7296930
Length=1940
Score = 32.7 bits (73), Expect = 0.35, Method: Composition-based stats.
Identities = 20/55 (36%), Positives = 31/55 (56%), Gaps = 1/55 (1%)
Query 1 AGA-AAAEISAAAGSSSGASSGASAGAAAGAAAAAAAAKSTRLSWSCGDTEGSLN 54
AGA + A +GS++GA G++AGA G+ AA + + SW G GS++
Sbjct 1806 AGAYPGSNAGAYSGSNAGAYPGSNAGAYPGSNAAWQSRTNAGSSWHSGPVAGSVS 1860
> CE26846
Length=467
Score = 30.0 bits (66), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 16/38 (42%), Positives = 20/38 (52%), Gaps = 2/38 (5%)
Query 129 PTIFKQRFCSSYATRNGTERCGRGNFCAFAHSREEVRA 166
P FK R C ++A G C G C FAH +E+RA
Sbjct 271 PHNFKTRLCMTHAA--GINPCALGARCKFAHGLKELRA 306
> ECU01g0830
Length=346
Score = 28.5 bits (62), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 21/71 (29%), Positives = 30/71 (42%), Gaps = 11/71 (15%)
Query 92 YRAEQCEEHFDSDSASLQCSKGDACEKSHNRHELLY---HPTIFKQRFCSSYATRNGTER 148
Y+ E C H + C GD C+ +H++ EL Y HP +K C ++
Sbjct 113 YKTEMCRSHTEIG----YCRYGDKCQFAHSKAELRYVQRHPK-YKTETCKTFWEEGS--- 164
Query 149 CGRGNFCAFAH 159
C G C F H
Sbjct 165 CPYGKRCCFIH 175
Lambda K H
0.318 0.126 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2526689620
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40