bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0621_orf3
Length=360
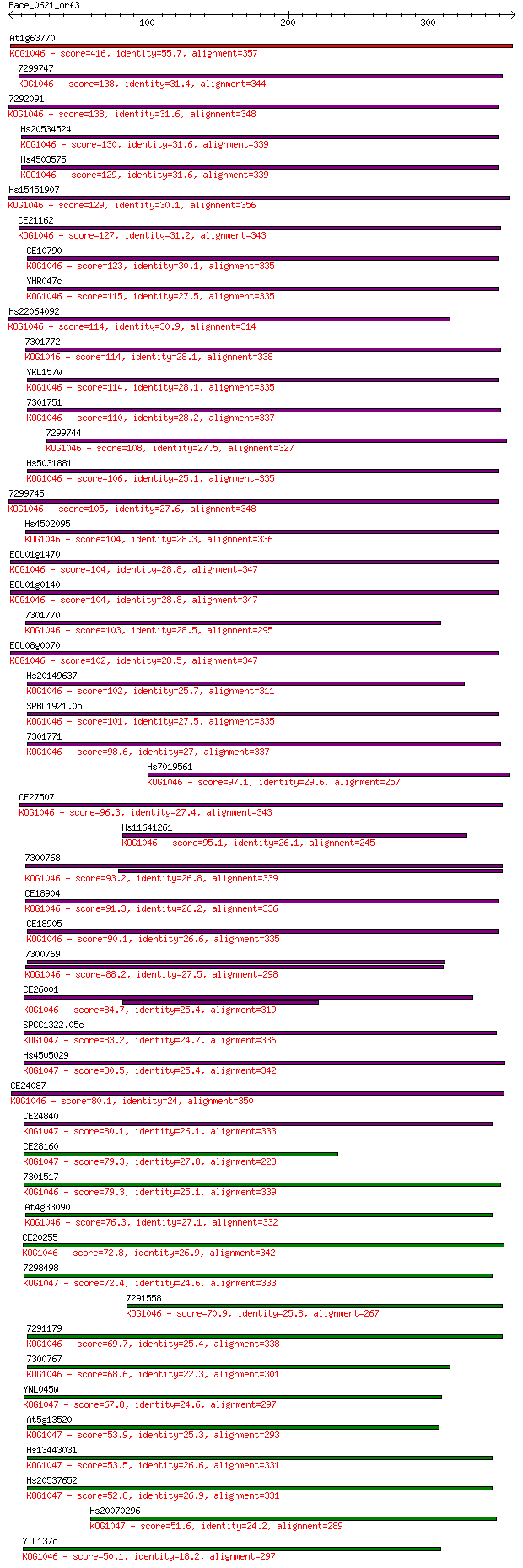
Score E
Sequences producing significant alignments: (Bits) Value
At1g63770 416 3e-116
7299747 138 2e-32
7292091 138 2e-32
Hs20534524 130 3e-30
Hs4503575 129 7e-30
Hs15451907 129 1e-29
CE21162 127 4e-29
CE10790 123 6e-28
YHR047c 115 1e-25
Hs22064092 114 4e-25
7301772 114 4e-25
YKL157w 114 4e-25
7301751 110 4e-24
7299744 108 2e-23
Hs5031881 106 1e-22
7299745 105 2e-22
Hs4502095 104 3e-22
ECU01g1470 104 3e-22
ECU01g0140 104 3e-22
7301770 103 8e-22
ECU08g0070 102 1e-21
Hs20149637 102 2e-21
SPBC1921.05 101 3e-21
7301771 98.6 2e-20
Hs7019561 97.1 5e-20
CE27507 96.3 8e-20
Hs11641261 95.1 2e-19
7300768 93.2 8e-19
CE18904 91.3 3e-18
CE18905 90.1 6e-18
7300769 88.2 2e-17
CE26001 84.7 3e-16
SPCC1322.05c 83.2 8e-16
Hs4505029 80.5 6e-15
CE24087 80.1 6e-15
CE24840 80.1 7e-15
CE28160 79.3 1e-14
7301517 79.3 1e-14
At4g33090 76.3 1e-13
CE20255 72.8 1e-12
7298498 72.4 2e-12
7291558 70.9 4e-12
7291179 69.7 1e-11
7300767 68.6 2e-11
YNL045w 67.8 3e-11
At5g13520 53.9 5e-07
Hs13443031 53.5 6e-07
Hs20537652 52.8 1e-06
Hs20070296 51.6 2e-06
YIL137c 50.1 7e-06
> At1g63770
Length=964
Score = 416 bits (1070), Expect = 3e-116, Method: Compositional matrix adjust.
Identities = 199/361 (55%), Positives = 257/361 (71%), Gaps = 7/361 (1%)
Query 2 LKGLYVSGSALVTQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVLLSNGNKTEEG 61
L+GLY S TQCEAEGFR+IT++ DRPD++AKY R+E +K YPVLLSNGN +G
Sbjct 187 LEGLYKSSGNFCTQCEAEGFRKITFYQDRPDIMAKYTCRVEGDKTLYPVLLSNGNLISQG 246
Query 62 LVHGSPDRHFAVFTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKL 121
+ G RH+A++ DP KP YLFA++AG+ + D FTT+SG +V+L +++ L K
Sbjct 247 DIEGG--RHYALWEDPFKKPCYLFALVAGQLVSRDDTFTTRSGRQVSLKIWTPAEDLPKT 304
Query 122 EWAMHSVKVSMKWEEEHFGREYDLDIFNVACVSDFNAGAMENKGLNIFNCTLLLADMQTS 181
AM+S+K +MKW+E+ FG EYDLD+FN+ V DFN GAMENK LNIFN L+LA +T+
Sbjct 305 AHAMYSLKAAMKWDEDVFGLEYDLDLFNIVAVPDFNMGAMENKSLNIFNSKLVLASPETA 364
Query 182 TDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGTVMSAGVQRIK 241
TD D+ +L V+GHEYFHNWTGNRVT RDWFQL+LKEGLTVFR+Q F + S V+RI
Sbjct 365 TDADYAAILGVIGHEYFHNWTGNRVTCRDWFQLSLKEGLTVFRDQEFSSDMGSRTVKRIA 424
Query 242 EVADLISRQFAEDDGPMAHPIRPESYMAMDN----FYTATVYDKGAEVVRIYHTLLGASG 297
+V+ L QF +D GPMAHP+RP SY+ + F + + GAEVVR+Y TLLG G
Sbjct 425 DVSKLRIYQFPQDAGPMAHPVRPHSYIKVYEKVWLFTNSVLLYAGAEVVRMYKTLLGTQG 484
Query 298 FRKGMDMYFERHDNTAATCDDFRAAMADANRINLDQMDRWYSQAGTPRLEVLRSEYKEDA 357
FRKG+D+YFERHD A TC+DF AAM DAN + +WYSQAGTP ++V+ S Y DA
Sbjct 485 FRKGIDLYFERHDEQAVTCEDFFAAMRDANNADFANFLQWYSQAGTPVVKVV-SSYNADA 543
Query 358 Q 358
+
Sbjct 544 R 544
> 7299747
Length=985
Score = 138 bits (348), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 108/357 (30%), Positives = 162/357 (45%), Gaps = 27/357 (7%)
Query 8 SGSALVTQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVLLSNGNKTEEGLVHGSP 67
S S + T+ E R+ D P + A++ + + +LSN E V G
Sbjct 267 SRSIISTKFEPTYARQAFPCFDEPALKAQFTITVARPSGDEYHVLSNMPVASE-YVDG-- 323
Query 68 DRHFAVFTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMHS 127
D F + P +YL A + +F + K TT G +AL VY+ PAQ+ K ++A+ +
Sbjct 324 DITEVTFAETVPMSTYLAAFVVSDF---QYKETTVEGTSIALKVYAPPAQVEKTQYALDT 380
Query 128 VKVSMKWEEEHFGREYDLDIFNVACVSDFNAGAMENKGLNIFNCTLLLADMQTSTDDDFE 187
M + +F Y L ++ + DF +GAMEN GL F T LL D TS+ + +
Sbjct 381 AAGVMAYYINYFNVSYALPKLDLVAIPDFVSGAMENWGLVTFRETALLYDESTSSSVNKQ 440
Query 188 RVLAVVGHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGTVMSAGVQRIKEVADLI 247
RV VV HE H W GN VT+ W L L EG F E GV+++ D+
Sbjct 441 RVAIVVAHELAHQWFGNLVTMNWWNDLWLNEGFASFLEY--------KGVKQMHPEWDMD 492
Query 248 SRQFAEDDGPM--------AHPI--RPESYMAMDNFYTATVYDKGAEVVRIYHTLLGASG 297
++ E+ P+ +HPI ES + ++ Y KGA +VR+ L+G
Sbjct 493 NQFVIEELHPVLTIDATLASHPIVKSIESPAEITEYFDTITYSKGAALVRMLENLVGEEK 552
Query 298 FRKGMDMYFERHDNTAATCDDFRAAMADANRINLDQ---MDRWYSQAGTPRLEVLRS 351
R Y RH + AT +D+ A+ + + D M W Q G P +EV +S
Sbjct 553 LRNATTRYLVRHIYSTATTEDYLTAVEEEEGLEFDVKQIMQTWTEQMGLPVVEVEKS 609
> 7292091
Length=866
Score = 138 bits (348), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 110/361 (30%), Positives = 168/361 (46%), Gaps = 22/361 (6%)
Query 1 KLKGLYVSGS---------ALVTQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVL 51
K+KG Y S A VTQ EA RR D P + A + + L K + V
Sbjct 111 KMKGFYRSKYFGPNGEERFAGVTQFEATDARRCFPCWDEPAIKATFDITLVVPKDR--VA 168
Query 52 LSNGNKTEEGLVHGSPDRHFAVFTDPHP-KPSYLFAILAGEFAAIRDKFTTKSGNEVALA 110
LSN +E + PD V D P +YL A++ GE+ D KS + V +
Sbjct 169 LSNMPVIKEDSL---PDGLRRVRFDRTPIMSTYLVAVVVGEY----DYVEGKSDDGVLVR 221
Query 111 VYSEPAQLHKLEWAMHSVKVSMKWEEEHFGREYDLDIFNVACVSDFNAGAMENKGLNIFN 170
V++ + + +A+ + + +++F Y L ++ +SDF+AGAMEN GL +
Sbjct 222 VFTPVGKREQGTFALEVATKVLPYYKDYFNIAYPLPKMDLIAISDFSAGAMENWGLVTYR 281
Query 171 CTLLLADMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMG 230
T +L D + ++ + + VGHE H W GN VT+ W L L EG F E L +
Sbjct 282 ETFVLVDPKNTSLMRKQSIALTVGHEIAHQWFGNLVTMEWWTHLWLNEGYASFVEFLCVH 341
Query 231 TVMSAGVQRIKEVADLISRQFAEDDGPMAHPIR-PESYMA-MDNFYTATVYDKGAEVVRI 288
+ + V D+ +R D +HPI P + + +D + Y+KGA V+R+
Sbjct 342 HLFPEYDIWTQFVTDMYTRALELDSLKNSHPIEVPVGHPSEIDEIFDEISYNKGASVIRM 401
Query 289 YHTLLGASGFRKGMDMYFERHDNTAATCDDFRAAMADANRINL-DQMDRWYSQAGTPRLE 347
H +G FRKGM++Y RH +D AA+ +A+ N+ D M W G P +
Sbjct 402 LHDYIGEDTFRKGMNLYLTRHQYGNTCTEDLWAALQEASSKNVSDVMTSWTQHKGFPVVS 461
Query 348 V 348
V
Sbjct 462 V 462
> Hs20534524
Length=957
Score = 130 bits (328), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 107/349 (30%), Positives = 160/349 (45%), Gaps = 25/349 (7%)
Query 10 SALVTQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVLLSNGNKTEEGLVHGSPDR 69
S + T E R+ D P+ A Y + + K +Y L SN +E V R
Sbjct 216 SIVATDHEPTDARKSFPCFDEPNKKATYTISITHPK-EYGAL-SNMPVAKEESVDDKWTR 273
Query 70 HFAVFTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMHSVK 129
F P +YL +F ++ K + SG L +Y +P Q H E+A + K
Sbjct 274 --TTFEKSVPMSTYLVCFAVHQFDSV--KRISNSGK--PLTIYVQPEQKHTAEYAANITK 327
Query 130 VSMKWEEEHFGREYDLDIFNVACVSDFNAGAMENKGLNIFNCTLLLADMQTSTDDDFERV 189
+ EE+F Y L + + DF GAMEN GL + T LL D + S + +RV
Sbjct 328 SVFDYFEEYFAMNYSLPKLDKIAIPDFGTGAMENWGLITYRETNLLYDPKESASSNQQRV 387
Query 190 LAVVGHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGTVMSAGVQRIKE---VADL 246
VV HE H W GN VT+ W L L EG F E F+G + ++++ + D+
Sbjct 388 ATVVAHELVHQWFGNIVTMDWWEDLWLNEGFASFFE--FLGVNHAETDWQMRDQMLLEDV 445
Query 247 ISRQFAEDDGPM-AHPI-----RPESYMAMDNFYTATVYDKGAEVVRIYHTLLGASGFRK 300
+ Q EDD M +HPI P+ ++ + Y KG+ ++R+ + F+K
Sbjct 446 LPVQ--EDDSLMSSHPIIVTVTTPDEITSV---FDGISYSKGSSILRMLEDWIKPENFQK 500
Query 301 GMDMYFERHDNTAATCDDFRAAMADANRINLDQ-MDRWYSQAGTPRLEV 348
G MY E++ A DF AA+ +A+R+ + + MD W Q G P L V
Sbjct 501 GCQMYLEKYQFKNAKTSDFWAALEEASRLPVKEVMDTWTRQMGYPVLNV 549
> Hs4503575
Length=957
Score = 129 bits (325), Expect = 7e-30, Method: Compositional matrix adjust.
Identities = 107/349 (30%), Positives = 159/349 (45%), Gaps = 25/349 (7%)
Query 10 SALVTQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVLLSNGNKTEEGLVHGSPDR 69
S T E R+ D P+ A Y + + K +Y L SN +E V R
Sbjct 216 SIAATDHEPTDARKSFPCFDEPNKKATYTISITHPK-EYGAL-SNMPVAKEESVDDKWTR 273
Query 70 HFAVFTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMHSVK 129
F P +YL +F ++ K + SG L +Y +P Q H E+A + K
Sbjct 274 --TTFEKSVPMSTYLVCFAVHQFDSV--KRISNSGK--PLTIYVQPEQKHTAEYAANITK 327
Query 130 VSMKWEEEHFGREYDLDIFNVACVSDFNAGAMENKGLNIFNCTLLLADMQTSTDDDFERV 189
+ EE+F Y L + + DF GAMEN GL + T LL D + S + +RV
Sbjct 328 SVFDYFEEYFAMNYSLPKLDKIAIPDFGTGAMENWGLITYRETNLLYDPKESASSNQQRV 387
Query 190 LAVVGHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGTVMSAGVQRIKE---VADL 246
VV HE H W GN VT+ W L L EG F E F+G + ++++ + D+
Sbjct 388 ATVVAHELVHQWFGNIVTMDWWEDLWLNEGFASFFE--FLGVNHAETDWQMRDQMLLEDV 445
Query 247 ISRQFAEDDGPM-AHPI-----RPESYMAMDNFYTATVYDKGAEVVRIYHTLLGASGFRK 300
+ Q EDD M +HPI P+ ++ + Y KG+ ++R+ + F+K
Sbjct 446 LPVQ--EDDSLMSSHPIIVTVTTPDEITSV---FDGISYSKGSSILRMLEDWIKPENFQK 500
Query 301 GMDMYFERHDNTAATCDDFRAAMADANRINLDQ-MDRWYSQAGTPRLEV 348
G MY E++ A DF AA+ +A+R+ + + MD W Q G P L V
Sbjct 501 GCQMYLEKYQFKNAKTSDFWAALEEASRLPVKEVMDTWTRQMGYPVLNV 549
> Hs15451907
Length=875
Score = 129 bits (323), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 107/376 (28%), Positives = 166/376 (44%), Gaps = 34/376 (9%)
Query 1 KLKGLYVSGS---------ALVTQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVL 51
K+KG Y S A VTQ EA RR D P + A + + L K + V
Sbjct 111 KMKGFYRSKYTTPSGEVRYAAVTQFEATDARRAFPCWDEPAIKATFDISLVVPKDR--VA 168
Query 52 LSNGNKTEEGLVHGSPDRHFAVFTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAV 111
LSN N + + F +YL A + GE+ D T+S + V + V
Sbjct 169 LSNMNVIDRKPYPDDENLVEVKFARTPVMSTYLVAFVVGEY----DFVETRSKDGVCVRV 224
Query 112 YSEPAQLHKLEWAMHSVKVSMKWEEEHFGREYDLDIFNVACVSDFNAGAMENKGLNIFNC 171
Y+ + + ++A+ ++ + +++F Y L ++ ++DF AGAMEN GL +
Sbjct 225 YTPVGKAEQGKFALEVAAKTLPFYKDYFNVPYPLPKIDLIAIADFAAGAMENWGLVTYRE 284
Query 172 TLLLADMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFM-- 229
T LL D + S + V VVGHE H W GN VT+ W L L EG + E L +
Sbjct 285 TALLIDPKNSCSSSRQWVALVVGHELAHQWFGNLVTMEWWTHLWLNEGFASWIEYLCVDH 344
Query 230 --------GTVMSAGVQRIKEVADLISRQFAEDDGPMAHPIRPESYMAMDNFYTATVYDK 281
+SA R +E+ L + E + HP +D + A Y K
Sbjct 345 CFPEYDIWTQFVSADYTRAQELDALDNSHPIEVS--VGHPSE------VDEIFDAISYSK 396
Query 282 GAEVVRIYHTLLGASGFRKGMDMYFERHDNTAATCDDFRAAMADANRINLDQ-MDRWYSQ 340
GA V+R+ H +G F+KGM+MY + A +D ++ +A+ + M+ W Q
Sbjct 397 GASVIRMLHDYIGDKDFKKGMNMYLTKFQQKNAATEDLWESLENASGKPIAAVMNTWTKQ 456
Query 341 AGTPRLEVLRSEYKED 356
G P + V + ++D
Sbjct 457 MGFPLIYVEAEQVEDD 472
> CE21162
Length=988
Score = 127 bits (318), Expect = 4e-29, Method: Compositional matrix adjust.
Identities = 107/361 (29%), Positives = 166/361 (45%), Gaps = 30/361 (8%)
Query 8 SGSALVTQCEAEGFRRITYFLDRPDVLAKYKVR-LEANKAKYPVLLSNGNKTEEGLVHGS 66
S A VTQ E RR+ D P A + V + NK V +SNG E+ + G
Sbjct 217 SKMAAVTQMEPVYARRMVPCFDEPAYKATWTVTVIHPNKT---VAVSNG--IEDKVEDGQ 271
Query 67 PDRHFAVFTDPHPK-PSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAM 125
P + F P P+ SYL AI EF ++ TTKSG V V+S P + + +A+
Sbjct 272 PGFIISTF-KPTPRMSSYLLAIFISEFE--YNEATTKSG--VRFRVWSRPEEKNSTMYAV 326
Query 126 HSVKVSMKWEEEHFGREYDLDIFNVACVSDFNAGAMENKGLNIFNCTLLLADMQTSTDDD 185
+ +++ E+++ + L ++ + DF+AGAMEN GL + + LL D + +
Sbjct 327 EAGVKCLEYYEKYYNISFPLPKQDMVALPDFSAGAMENWGLITYRESALLYDPRIYSGSQ 386
Query 186 FERVLAVVGHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGT-VMSAGVQRIKE-- 242
RV V+ HE H W GN VT++ W L L EG E ++GT +S G R++E
Sbjct 387 KRRVAVVIAHELAHQWFGNLVTLKWWNDLWLNEGFATLVE--YLGTDEISDGNMRMREWF 444
Query 243 VADLISRQFAEDDGPMAHPI--RPESYMAMDNFYTATVYDKGAEVVRIYHTLLGASGFRK 300
D + A D HP+ + + M + + + + YDKG V+ + +G F
Sbjct 445 TMDALWSALAADSVASTHPLTFKIDKAMEVLDSFDSVTYDKGGAVLAMVRKTIGEENFNT 504
Query 301 GMDMYFERHDNTAATCDDFRAAMAD----------ANRINLDQ-MDRWYSQAGTPRLEVL 349
G++ Y RH A + A+ + ++N+ + MD W Q G P L
Sbjct 505 GINHYLTRHQFDNADAGNLLTALGEKIPDSVMGPKGVKLNISEFMDPWTKQLGYPLLNAS 564
Query 350 R 350
R
Sbjct 565 R 565
> CE10790
Length=884
Score = 123 bits (308), Expect = 6e-28, Method: Compositional matrix adjust.
Identities = 101/338 (29%), Positives = 149/338 (44%), Gaps = 10/338 (2%)
Query 14 TQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVLLSNGNKTEEGLVHGSPDRHFAV 73
TQ E+ R D P A + V LE + LSN N E +
Sbjct 139 TQFESTYARYAFPCFDEPIYKATFDVTLEVEN--HLTALSNMNVISETPTADGKRKAVTF 196
Query 74 FTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMHSVKVSMK 133
T P SYL A GE I + TKSG V + VY+ P + + ++++ +
Sbjct 197 ATSPK-MSSYLVAFAVGELEYISAQ--TKSG--VEMRVYTVPGKKEQGQYSLDLSVKCID 251
Query 134 WEEEHFGREYDLDIFNVACVSDFNAGAMENKGLNIFNCTLLLADMQTSTDDDFERVLAVV 193
W E F +Y L ++ + DF+ GAMEN GL + LL D ++ RV VV
Sbjct 252 WYNEWFDIKYPLPKCDLIAIPDFSMGAMENWGLVTYREIALLVDPGVTSTRQKSRVALVV 311
Query 194 GHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGTVMSAGVQRIKEVADLISRQFAE 253
HE H W GN VT++ W L LKEG F E +F+G + + D ++
Sbjct 312 AHELAHLWFGNLVTMKWWTDLWLKEGFASFMEYMFVGANCPEFKIWLHFLNDELASGMGL 371
Query 254 DDGPMAHPIRPE--SYMAMDNFYTATVYDKGAEVVRIYHTLLGASGFRKGMDMYFERHDN 311
D +HPI E + +D Y + Y K V R+ L F+KG+ +Y +R
Sbjct 372 DALRNSHPIEVEIDNPNELDEIYDSITYAKSNSVNRMLCYYLSEPVFQKGLRLYLKRFQY 431
Query 312 TAATCDDFRAAMADANRINLDQ-MDRWYSQAGTPRLEV 348
+ A D A+++A+ N+++ M W Q G P L+V
Sbjct 432 SNAVTQDLWTALSEASGQNVNELMSGWTQQMGFPVLKV 469
> YHR047c
Length=856
Score = 115 bits (289), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 92/338 (27%), Positives = 147/338 (43%), Gaps = 13/338 (3%)
Query 14 TQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVLLSNGNKTEEGLVHGSPDRHFAV 73
TQ EA RR D P++ A + V L + + LSN + E + G + +
Sbjct 129 TQMEATDARRAFPCFDEPNLKATFAVTLVSES--FLTHLSNMDVRNETIKEG---KKYTT 183
Query 74 FTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMHSVKVSMK 133
F +YL A + + ++ + + + VYS P ++A + +++
Sbjct 184 FNTTPKMSTYLVAFIVADL-----RYVESNNFRIPVRVYSTPGDEKFGQFAANLAARTLR 238
Query 134 WEEEHFGREYDLDIFNVACVSDFNAGAMENKGLNIFNCTLLLADMQTSTDDDFERVLAVV 193
+ E+ F EY L ++ V +F+AGAMEN GL + LL D++ S+ D +RV V+
Sbjct 239 FFEDTFNIEYPLPKMDMVAVHEFSAGAMENWGLVTYRVIDLLLDIENSSLDRIQRVAEVI 298
Query 194 GHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGTVMSAGVQRIKEVADLISRQFAE 253
HE H W GN VT+ W L L EG + + V D + R
Sbjct 299 QHELAHQWFGNLVTMDWWEGLWLNEGFATWMSWYSCNKFQPEWKVWEQYVTDNLQRALNL 358
Query 254 DDGPMAHPIRPESYMA--MDNFYTATVYDKGAEVVRIYHTLLGASGFRKGMDMYFERHDN 311
D +HPI A ++ + A Y KG+ ++R+ LG F KG+ Y +
Sbjct 359 DSLRSSHPIEVPVNNADEINQIFDAISYSKGSSLLRMISKWLGEETFIKGVSQYLNKFKY 418
Query 312 TAATCDDFRAAMADANRINL-DQMDRWYSQAGTPRLEV 348
A D A+ADA+ ++ M+ W + G P L V
Sbjct 419 GNAKTGDLWDALADASGKDVCSVMNIWTKRVGFPVLSV 456
> Hs22064092
Length=478
Score = 114 bits (284), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 97/330 (29%), Positives = 144/330 (43%), Gaps = 25/330 (7%)
Query 1 KLKGLYVSGS---------ALVTQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVL 51
K+KG Y S A VTQ EA RR D + A + + L K + V
Sbjct 155 KMKGFYRSKYTTPSGEVRYAAVTQFEATDARRAFPCWDERAIKATFDISLVVPKDR--VA 212
Query 52 LSNGNKTEEGLVHGSPDRHFAVFTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAV 111
LSN N + + F +YL A + GE+ D T+S + V + V
Sbjct 213 LSNMNVIDRKPYPDDENLVEVKFARTPVTSTYLVAFVVGEY----DFVETRSKDGVCVCV 268
Query 112 YSEPAQLHKLEWAMHSVKVSMKWEEEHFGREYDLDIFNVACVSDFNAGAMENKGLNIFNC 171
Y+ + + ++A+ ++ + +++F Y L ++ ++DF AGAMEN L +
Sbjct 269 YTPVGKAEQGKFALEVAAKTLPFYKDYFNVPYPLPKIDLIAIADFAAGAMENWDLVTYRE 328
Query 172 TLLLADMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGT 231
T LL D + S + V VVGHE H W GN VT+ W L L EG + E L +
Sbjct 329 TALLIDPKNSCSSSRQWVALVVGHELAHQWFGNLVTMEWWTHLRLNEGFASWIEYLCVDH 388
Query 232 VMSAGVQRIKEVADLISRQFAEDDGPMAHPIR-----PESYMAMDNFYTATVYDKGAEVV 286
+ V+ +R D +HPI P +D + A Y KGA V+
Sbjct 389 CFPEYDIWTQFVSADYTRAQELDALDNSHPIEVSVGHPSE---VDEIFDAISYSKGASVI 445
Query 287 RIYHTLLGASGFRKGMDMYFER--HDNTAA 314
R+ H +G F+KGM+MY + N AA
Sbjct 446 RMLHDYIGDKDFKKGMNMYLTKFQQKNAAA 475
> 7301772
Length=932
Score = 114 bits (284), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 95/351 (27%), Positives = 149/351 (42%), Gaps = 21/351 (5%)
Query 13 VTQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVLLSNGNKTEEGLVHGSPDRHFA 72
VTQ E R D PD A + V L +K KY + SN + E D +
Sbjct 166 VTQFEPASARLAFPCFDEPDFKAPFVVTLGYHK-KYTAI-SNMPEKETKPHETLADYIWC 223
Query 73 VFTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMHSVKVSM 132
F + P +YL A +F+ N ++ P + + ++A +
Sbjct 224 EFQESVPMSTYLVAYSVNDFSHKPSTLP----NSALFRTWARPNAIDQCDYAAQFGPKVL 279
Query 133 KWEEEHFGREYDLDIFNVACVSDFNAGAMENKGLNIFNCTLLLADMQTSTDDDFERVLAV 192
++ E+ FG ++ L + V DF+AGAMEN GL + LL S+ D +RV +V
Sbjct 280 QYYEQFFGIKFPLPKIDQIAVPDFSAGAMENWGLVTYREIALLYSAAHSSLADKQRVASV 339
Query 193 VGHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGTV----MSAGVQRIKEVADLIS 248
V HE H W GN VT++ W L L EG + L + + S + + + +
Sbjct 340 VAHELAHQWFGNLVTMKWWTDLWLNEGFATYVASLGVENINPEWRSMEQESLSNLLTIFR 399
Query 249 RQFAEDDGPMAHPIRPESYMAMDNFYTATVYDKGAEVVRIYHTLLGASGFRKGMDMYFER 308
R E P++ PI+ S ++ + Y KG+ V+R+ H LG FR G+ Y ++
Sbjct 400 RDALESSHPISRPIQMVSEISES--FDQISYQKGSTVLRMMHLFLGEESFRSGLQAYLQK 457
Query 309 HDNTAATCDDFRAAMADANRI--NLDQ-------MDRWYSQAGTPRLEVLR 350
A D+ ++ A +L + MD W Q G P + V R
Sbjct 458 FSYKNAEQDNLWESLTQAAHKYRSLPKSYDIKSIMDSWTLQTGYPVINVTR 508
> YKL157w
Length=935
Score = 114 bits (284), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 94/340 (27%), Positives = 144/340 (42%), Gaps = 17/340 (5%)
Query 14 TQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVL--LSNGNKTEEGLVHGSPDRHF 71
TQ E RR D P++ A + + L ++ P L LSN + E + G +
Sbjct 225 TQMEPTDARRAFPCFDEPNLKASFAITLVSD----PSLTHLSNMDVKNEYVKDG---KKV 277
Query 72 AVFTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMHSVKVS 131
+F +YL A + E K+ + + VY+ P ++A +
Sbjct 278 TLFNTTPKMSTYLVAFIVAEL-----KYVESKNFRIPVRVYATPGNEKHGQFAADLTAKT 332
Query 132 MKWEEEHFGREYDLDIFNVACVSDFNAGAMENKGLNIFNCTLLLADMQTSTDDDFERVLA 191
+ + E+ FG +Y L + V +F+AGAMEN GL + LL D ST D +RV
Sbjct 333 LAFFEKTFGIQYPLPKMDNVAVHEFSAGAMENWGLVTYRVVDLLLDKDNSTLDRIQRVAE 392
Query 192 VVGHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGTVMSAGVQRIKEVADLISRQF 251
VV HE H W GN VT+ W L L EG + + V D +
Sbjct 393 VVQHELAHQWFGNLVTMDWWEGLWLNEGFATWMSWYSCNEFQPEWKVWEQYVTDTLQHAL 452
Query 252 AEDDGPMAHPIRPESYMA--MDNFYTATVYDKGAEVVRIYHTLLGASGFRKGMDMYFERH 309
+ D +HPI A ++ + A Y KGA ++R+ LG F KG+ Y +
Sbjct 453 SLDSLRSSHPIEVPVKKADEINQIFDAISYSKGASLLRMISKWLGEETFIKGVSQYLNKF 512
Query 310 DNTAATCDDFRAAMADANRINLDQ-MDRWYSQAGTPRLEV 348
A +D A+ADA+ ++ M+ W + G P + V
Sbjct 513 KYGNAKTEDLWDALADASGKDVRSVMNIWTKKVGFPVISV 552
> 7301751
Length=1017
Score = 110 bits (276), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 95/353 (26%), Positives = 151/353 (42%), Gaps = 32/353 (9%)
Query 14 TQCEAEGFRRITYFLDRPDVLAKYKVRLEANK-----AKYPVLLSNGNKTEEGLVHGSPD 68
TQ +A RR D P + A + + + + + P++ SN + T V
Sbjct 264 TQFQATDARRAFPCFDEPALKANFTLHIARPRNMTTISNMPIVSSNDHATMPSYVWD--- 320
Query 69 RHFAVFTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMHSV 128
HFA + P +YL A +F I SGN AV++ + E+A+
Sbjct 321 -HFA---ESLPMSTYLVAYAISDFTHI------SSGN---FAVWARADAIKSAEYALSVG 367
Query 129 KVSMKWEEEHFGREYDLDIFNVACVSDFNAGAMENKGLNIFNCTLLLADMQTSTDDDFER 188
+ + ++ F + L ++ + +F AGAMEN GL F T +L D +T ++ +R
Sbjct 368 PRILTFLQDFFNVTFPLPKIDMIALPEFQAGAMENWGLITFRETAMLYDPGVATANNKQR 427
Query 189 VLAVVGHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGTVMSAGVQRIKEVADLIS 248
V +VVGHE H W GN VT W + L EG + E L V Q + V + +
Sbjct 428 VASVVGHELAHQWFGNLVTPSWWSDIWLNEGFASYMEYLTADAVAPEWKQLDQFVVNELQ 487
Query 249 RQFAEDDGPMAHPIRPESY--MAMDNFYTATVYDKGAEVVRIYHTLLGASGFRKGMDMYF 306
F D +H I E + + + Y KG+ ++R+ L FR+G+ Y
Sbjct 488 AVFQLDALSTSHKISHEVFNPQEISEIFDRISYAKGSTIIRMMAHFLTNPIFRRGLSKYL 547
Query 307 ERHDNTAATCDDF---------RAAMADANRINLDQMDRWYSQAGTPRLEVLR 350
+ +AT DD + + D +R + MD W Q G P ++V R
Sbjct 548 QEMAYNSATQDDLWHFLTVEAKSSGLLDDSRSVKEIMDTWTLQTGYPVVKVSR 600
> 7299744
Length=988
Score = 108 bits (270), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 90/339 (26%), Positives = 157/339 (46%), Gaps = 25/339 (7%)
Query 28 LDRPDVLAKYKVRLEANKAKYPVLLSNGNKTEEGLVHGSPDRHFAVFTDPHPKPSYLFAI 87
D P + A++ + L + LSN N + + G+ F P +YL
Sbjct 261 FDEPALKAEFTITLVHPSGEDYHALSNMN-VDSSVSQGAFQE--VTFAKSVPMSTYLACF 317
Query 88 LAGEFAAIRDKFTTKS-GNEVALAVYSEPAQLHKLEWAMHSVKVSMKWEEEHFGREYDLD 146
+ +FA + TK G +++VY+ P QL K++ A+ K +++ ++F Y L
Sbjct 318 IVSDFAYKQVSIDTKGIGETFSMSVYATPEQLDKVDLAVTIGKGVIEYYIDYFQIAYPLP 377
Query 147 IFNVACVSDFNAGAMENKGLNIFNCTLLLADMQTSTDDDFERVLAVVGHEYFHNWTGNRV 206
++A + DF +GAME+ GL + T LL D TS+ + +R+ +V+ HE+ H W GN V
Sbjct 378 KLDMAAIPDFVSGAMEHWGLVTYRETSLLYDEATSSATNKQRIASVIAHEFAHMWFGNLV 437
Query 207 TVRDWFQLTLKEGLTVFREQLFMGTVMSAGVQRIKEVADLISRQFAEDDGPM-AHPI--R 263
T+ W L L EG F E L + V ++ +++ DG + +HPI
Sbjct 438 TMNWWNDLWLNEGFASFVEYLGVDAVYPE--WKMASAPSVLTL-----DGTLGSHPIIQT 490
Query 264 PESYMAMDNFYTATVYDKGAEVVRIYHTLLGASGFRKGMDMYFERHDNTAATCDDFRAAM 323
E+ + + Y KG+ +VR+ LG + FR+ + Y + + A +F
Sbjct 491 VENPDQITEIFDTITYSKGSSLVRMLEDFLGETTFRQAVTNYLNEYKYSTAETGNF---F 547
Query 324 ADANRINL-----DQMDRWYSQAGTPRLEVLR---SEYK 354
+ +++ L + M W Q G P + + + +EYK
Sbjct 548 TEIDKLELGYNVTEIMLTWTVQMGLPVVTIEKVSDTEYK 586
> Hs5031881
Length=1025
Score = 106 bits (264), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 84/344 (24%), Positives = 150/344 (43%), Gaps = 23/344 (6%)
Query 14 TQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVLLSNGNKT----EEGLVHGSPDR 69
TQ E R D P A + +++ ++ +Y L + K+ ++GLV
Sbjct 292 TQFEPLAARSAFPCFDEPAFKATFIIKIIRDE-QYTALSNMPKKSSVVLDDGLVQDE--- 347
Query 70 HFAVFTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMHSVK 129
F++ +YL A + GE K ++ N +++Y+ P + ++ +A+ +
Sbjct 348 ----FSESVKMSTYLVAFIVGEM-----KNLSQDVNGTLVSIYAVPENIGQVHYALETTV 398
Query 130 VSMKWEEEHFGREYDLDIFNVACVSDFNAGAMENKGLNIFNCTLLLADMQTSTDDDFERV 189
+++ + +F +Y L ++ + DF AGAMEN GL F LL D TS+ D + V
Sbjct 399 KLLEFFQNYFEIQYPLKKLDLVAIPDFEAGAMENWGLLTFREETLLYDSNTSSMADRKLV 458
Query 190 LAVVGHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGTVMSAGVQRIKEVADLISR 249
++ HE H W GN VT++ W L L EG F E + + + ++ D +
Sbjct 459 TKIIAHELAHQWFGNLVTMKWWNDLWLNEGFATFMEYFSLEKIFKE-LSSYEDFLDARFK 517
Query 250 QFAEDDGPMAHPIRP--ESYMAMDNFYTATVYDKGAEVVRIYHTLLGASGFRKGMDMYFE 307
+D +HPI +S ++ + + Y KG+ ++ + T L F+ + +Y
Sbjct 518 TMKKDSLNSSHPISSSVQSSEQIEEMFDSLSYFKGSSLLLMLKTYLSEDVFQHAVVLYLH 577
Query 308 RHDNTAATCDDFRAAMADANRINLD---QMDRWYSQAGTPRLEV 348
H + DD + + LD M W Q G P + V
Sbjct 578 NHSYASIQSDDLWDSFNEVTNQTLDVKRMMKTWTLQKGFPLVTV 621
> 7299745
Length=913
Score = 105 bits (261), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 96/368 (26%), Positives = 161/368 (43%), Gaps = 30/368 (8%)
Query 1 KLKGLYVSG---------SALVTQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVL 51
KL GLY S + T+ E R+ D P + A + + + Y +
Sbjct 165 KLVGLYSSTYLNEAGATRTISTTKFEPTYARQAFPCFDEPAMKATFAITVVHPSGSYHAV 224
Query 52 LSNGNKTEEGLVHGSPDRHFAVFTDPHPKPSYLFAILAGEFAAIRDKFTTKS---GNEVA 108
SN +TE + D A+F +YL I+ +FA+ T K+ G + +
Sbjct 225 -SNMQQTESNYLG---DYTEAIFETSVSMSTYLVCIIVSDFAS--QSTTVKANGIGEDFS 278
Query 109 LAVYSEPAQLHKLEWAMHSVKVSMKWEEEHFGREYDLDIFNVACVSDFNAGAMENKGLNI 168
+ Y+ Q++K+E+A+ + ++ +++ Y L ++A + DF +GAME+ GL
Sbjct 279 MQAYATSHQINKVEFALEFGQAVTEYYIQYYKVPYPLTKLDMAAIPDFASGAMEHWGLVT 338
Query 169 FNCTLLLADMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLF 228
+ T LL D S+ + + + + HE H W GN VT++ W L L EG + +
Sbjct 339 YRETALLYDPSYSSTANKQSIAGTLAHEIAHQWFGNLVTMKWWNDLWLNEGFARYMQYKG 398
Query 229 MGTVMS--AGVQRIKEVADLISRQFAEDDGPMAHPI--RPESYMAMDNFYTATVYDKGAE 284
+ V +++ + VA + D +HPI + ES + + Y+KG
Sbjct 399 VNAVHPDWGMLEQFQIVA--LQPVLLYDAKLSSHPIVQKVESPDEITAIFDTISYEKGGS 456
Query 285 VVRIYHTLLGASGFRKGMDMYFERHDNTAATCDDF----RAAMADANRINLDQMDRWYSQ 340
V+R+ TL+GA F + + Y +H DDF A + D + L M W Q
Sbjct 457 VIRMLETLVGAEKFEEAVTNYLVKHQFNNTVTDDFLTEVEAVVTDLDIKKL--MLTWTEQ 514
Query 341 AGTPRLEV 348
G P L V
Sbjct 515 MGYPVLNV 522
> Hs4502095
Length=967
Score = 104 bits (260), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 95/356 (26%), Positives = 154/356 (43%), Gaps = 30/356 (8%)
Query 13 VTQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVLLSNGNKTEEGLVHGSPDRHFA 72
TQ +A R+ D P + A++ + L K L + N +G P+
Sbjct 209 TTQMQAADARKSFPCFDEPAMKAEFNITLIHPKD----LTALSNMLPKGPSTPLPEDPNW 264
Query 73 VFTDPHPKP---SYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKL--EWAMHS 127
T+ H P +YL A + EF D ++ N V + +++ P+ + ++A++
Sbjct 265 NVTEFHTTPKMSTYLLAFIVSEF----DYVEKQASNGVLIRIWARPSAIAAGHGDYALNV 320
Query 128 VKVSMKWEEEHFGREYDLDIFNVACVSDFNAGAMENKGLNIFNCTLLLADMQTSTDDDFE 187
+ + H+ Y L + + DFNAGAMEN GL + LL D +S+ + E
Sbjct 321 TGPILNFFAGHYDTPYPLPKSDQIGLPDFNAGAMENWGLVTYRENSLLFDPLSSSSSNKE 380
Query 188 RVLAVVGHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGTVMSAGVQRIKEVADL- 246
RV+ V+ HE H W GN VT+ W L L EG + E ++G + +K++ L
Sbjct 381 RVVTVIAHELAHQWFGNLVTIEWWNDLWLNEGFASYVE--YLGADYAEPTWNLKDLMVLN 438
Query 247 -ISRQFAEDDGPMAHPIR-PESYM----AMDNFYTATVYDKGAEVVRIYHTLLGASGFRK 300
+ R A D +HP+ P S + + + A Y KGA V+R+ + L F++
Sbjct 439 DVYRVMAVDALASSHPLSTPASEINTPAQISELFDAISYSKGASVLRMLSSFLSEDVFKQ 498
Query 301 GMDMY---FERHDNTAATCDDFRAAMADANRINL-----DQMDRWYSQAGTPRLEV 348
G+ Y F + D + I L D M+RW Q G P + V
Sbjct 499 GLASYLHTFAYQNTIYLNLWDHLQEAVNNRSIQLPTTVRDIMNRWTLQMGFPVITV 554
> ECU01g1470
Length=864
Score = 104 bits (260), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 100/361 (27%), Positives = 160/361 (44%), Gaps = 22/361 (6%)
Query 2 LKGLYVSGS---ALVTQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVLLSNGNKT 58
L GLY SG T E RR D+PD+ A +K+ ++A +K+ VL + +
Sbjct 131 LVGLYKSGGPKEVYSTHFEPTDARRAFPCFDQPDMKATFKISIDAG-SKFTVLANT--QA 187
Query 59 EEGLVHGSPDRHFAVFTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQL 118
L DR F + +YL A + GE + I D +K G V L VY + +++
Sbjct 188 IPSLREEYGDRKIEYFEETCKMSTYLVAFVVGELSYIED--WSKDG--VRLRVYGDSSEV 243
Query 119 HKLEWAMHSVKVSMKWEEEHFGREYDLD-----IFNVACVSDFNAGAMENKGLNIFNCTL 173
+ + K +++ E+FG Y+ ++ + +F++GAMEN GL F
Sbjct 244 EWGRYGLEVGKRCLEYFSEYFGVGYEFPRAGSAKIDMVGIPNFSSGAMENWGLITFRRES 303
Query 174 LLADMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTLKEGLTV---FREQLFMG 230
LL S +D + V V HE H W GN VT+ W L L EG F+ +G
Sbjct 304 LLYVPGKSNVEDMKNVAGTVCHELGHMWFGNLVTMSWWDDLWLNEGFATWVSFKGMENIG 363
Query 231 TVMSAGVQRIKEVADLISRQFAEDDGPMAHPIRPESY--MAMDNFYTATVYDKGAEVVRI 288
+V+S V + +++ R +D +H IR + + + Y KGA V+R+
Sbjct 364 SVVSWDVWGEFVLWNVV-RGMVDDGLGKSHQIRMNVTDPGEIGEIFDSISYCKGASVIRM 422
Query 289 YHTLLGASGFRKGMDMYFERHDNTAATCDDFRAAMADANRINLDQM-DRWYSQAGTPRLE 347
+G S F G+ Y + H A+ + ++ +M + W SQAG P +
Sbjct 423 IERYVGESVFMLGIRRYIKEHMYGNGNAMSLWKAIGEEYGEDISEMVEGWISQAGYPVVS 482
Query 348 V 348
V
Sbjct 483 V 483
> ECU01g0140
Length=864
Score = 104 bits (260), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 100/361 (27%), Positives = 160/361 (44%), Gaps = 22/361 (6%)
Query 2 LKGLYVSGS---ALVTQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVLLSNGNKT 58
L GLY SG T E RR D+PD+ A +K+ ++A +K+ VL + +
Sbjct 131 LVGLYKSGGPKEVYSTHFEPTDARRAFPCFDQPDMKATFKISIDAG-SKFTVLANT--QA 187
Query 59 EEGLVHGSPDRHFAVFTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQL 118
L DR F + +YL A + GE + I D +K G V L VY + +++
Sbjct 188 IPSLREEYGDRKIEYFEETCKMSTYLVAFVVGELSYIED--WSKDG--VRLRVYGDSSEV 243
Query 119 HKLEWAMHSVKVSMKWEEEHFGREYDLD-----IFNVACVSDFNAGAMENKGLNIFNCTL 173
+ + K +++ E+FG Y+ ++ + +F++GAMEN GL F
Sbjct 244 EWGRYGLEVGKRCLEYFSEYFGVGYEFPRAGSAKIDMVGIPNFSSGAMENWGLITFRRES 303
Query 174 LLADMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTLKEGLTV---FREQLFMG 230
LL S +D + V V HE H W GN VT+ W L L EG F+ +G
Sbjct 304 LLYVPGKSNVEDMKNVAGTVCHELGHMWFGNLVTMSWWDDLWLNEGFATWVSFKGMENIG 363
Query 231 TVMSAGVQRIKEVADLISRQFAEDDGPMAHPIRPESY--MAMDNFYTATVYDKGAEVVRI 288
+V+S V + +++ R +D +H IR + + + Y KGA V+R+
Sbjct 364 SVVSWDVWGEFVLWNVV-RGMVDDGLGKSHQIRMNVTDPGEIGEIFDSISYCKGASVIRM 422
Query 289 YHTLLGASGFRKGMDMYFERHDNTAATCDDFRAAMADANRINLDQM-DRWYSQAGTPRLE 347
+G S F G+ Y + H A+ + ++ +M + W SQAG P +
Sbjct 423 IERYVGESVFMLGIRRYIKEHMYGNGNAMSLWKAIGEEYGEDISEMVEGWISQAGYPVVS 482
Query 348 V 348
V
Sbjct 483 V 483
> 7301770
Length=814
Score = 103 bits (256), Expect = 8e-22, Method: Compositional matrix adjust.
Identities = 84/298 (28%), Positives = 132/298 (44%), Gaps = 10/298 (3%)
Query 13 VTQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVLLSNGNKTEEGLVHGSPDRHFA 72
VTQ E R D P A + + L +K KY LSN E PD +
Sbjct 159 VTQFEPAAARLAFPCFDEPGYKASFAITLGYHK-KY-TGLSNMPVNETRPHESIPDYVWT 216
Query 73 VFTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMHSVKVSM 132
F + P +YL A +F+ N ++ P + + ++A +
Sbjct 217 SFEESLPMSTYLVAYSLNDFSHKPSTLP----NGTLFRTWARPNAIDQCDYAAELGPKVL 272
Query 133 KWEEEHFGREYDLDIFNVACVSDFNAGAMENKGLNIFNCTLLLADMQTSTDDDFERVLAV 192
K+ EE FG ++ L + V DF+AGAMEN GL F + LL + S+ + + +
Sbjct 273 KYYEELFGIKFPLPKVDQIAVPDFSAGAMENWGLVTFAESTLLYSPEYSSQEAKQETANI 332
Query 193 VGHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGTVMSAG-VQRIKEVADLISRQF 251
V HE H W GN VT++ W L L EG + L + + +++ +++L++ F
Sbjct 333 VAHELAHQWFGNLVTMKWWTDLWLNEGFATYVASLCVEDIHPQWRTLQLESMSNLLTI-F 391
Query 252 AEDDGPMAHPI-RP-ESYMAMDNFYTATVYDKGAEVVRIYHTLLGASGFRKGMDMYFE 307
+D +HPI RP E + + Y KG+ V+R+ H LG FR G+ + E
Sbjct 392 RKDSLESSHPISRPIEMTSQISESFDEISYQKGSSVIRMMHLFLGEEAFRTGLKLVLE 449
> ECU08g0070
Length=864
Score = 102 bits (254), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 99/361 (27%), Positives = 160/361 (44%), Gaps = 22/361 (6%)
Query 2 LKGLYVSGS---ALVTQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVLLSNGNKT 58
L GLY SG T E R + D+PD+ A +K+ ++A +K+ VL + +
Sbjct 131 LVGLYKSGGPKEVYSTHFEPTDARWVFPCFDQPDMKATFKISIDAG-SKFTVLANT--QA 187
Query 59 EEGLVHGSPDRHFAVFTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQL 118
L DR F + +YL A + GE + I D +K G V L VY + +++
Sbjct 188 IPSLREEYGDRKIEYFEETCKMSTYLVAFVVGELSYIED--WSKDG--VRLRVYGDSSEV 243
Query 119 HKLEWAMHSVKVSMKWEEEHFGREYDLD-----IFNVACVSDFNAGAMENKGLNIFNCTL 173
+ + K +++ E+FG Y+ ++ + +F++GAMEN GL F
Sbjct 244 EWGRYGLEVGKRCLEYFSEYFGVGYEFPRAGSAKIDMVGIPNFSSGAMENWGLITFRRES 303
Query 174 LLADMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTLKEGLTV---FREQLFMG 230
LL S +D + V V HE H W GN VT+ W L L EG F+ +G
Sbjct 304 LLYVPGKSNVEDMKNVAETVCHELGHMWFGNLVTMSWWDDLWLNEGFATWVSFKGMENIG 363
Query 231 TVMSAGVQRIKEVADLISRQFAEDDGPMAHPIRPESY--MAMDNFYTATVYDKGAEVVRI 288
+V+S V + +++ R +D +H IR + + + Y KGA V+R+
Sbjct 364 SVVSWDVWGEFVLWNVV-RGMVDDGLGKSHQIRMNVTDPGEIGEIFDSISYCKGASVIRM 422
Query 289 YHTLLGASGFRKGMDMYFERHDNTAATCDDFRAAMADANRINLDQM-DRWYSQAGTPRLE 347
+G S F G+ Y + H A+ + ++ +M + W SQAG P +
Sbjct 423 IERYVGESVFMLGIRRYIKEHMYGNGNAMSLWKAIGEEYGEDISEMVEGWISQAGYPVVS 482
Query 348 V 348
V
Sbjct 483 V 483
> Hs20149637
Length=948
Score = 102 bits (253), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 80/317 (25%), Positives = 138/317 (43%), Gaps = 19/317 (5%)
Query 14 TQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVL---LSNGNKTEEGLVHGSPDRH 70
TQ E R D P A + +++ + L EGL+ + H
Sbjct 180 TQFEPTAARMAFPCFDEPAFKASFSIKIRREPRHLAISNMPLVKSVTVAEGLI----EDH 235
Query 71 FAVFTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMHSVKV 130
F V +YL A + +F ++ TKSG V ++VY+ P ++++ ++A+ +
Sbjct 236 FDVTVK---MSTYLVAFIISDFESVSK--ITKSG--VKVSVYAVPDKINQADYALDAAVT 288
Query 131 SMKWEEEHFGREYDLDIFNVACVSDFNAGAMENKGLNIFNCTLLLADMQTSTDDDFERVL 190
+++ E++F Y L ++A + DF +GAMEN GL + + LL D + S+ +
Sbjct 289 LLEFYEDYFSIPYPLPKQDLAAIPDFQSGAMENWGLTTYRESALLFDAEKSSASSKLGIT 348
Query 191 AVVGHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGTV---MSAGVQRIKEVADLI 247
V HE H W GN VT+ W L L EG F E + + + G + D +
Sbjct 349 VTVAHELAHQWFGNLVTMEWWNDLWLNEGFAKFMEFVSVSVTHPELKVGDYFFGKCFDAM 408
Query 248 SRQFAEDDGPMAHPIRPESYMAMDNFYTATVYDKGAEVVRIYHTLLGASGFRKGMDMYFE 307
P++ P+ E+ + + YDKGA ++ + L A F+ G+ Y +
Sbjct 409 EVDALNSSHPVSTPV--ENPAQIREMFDDVSYDKGACILNMLREYLSADAFKSGIVQYLQ 466
Query 308 RHDNTAATCDDFRAAMA 324
+H +D +MA
Sbjct 467 KHSYKNTKNEDLWDSMA 483
> SPBC1921.05
Length=882
Score = 101 bits (251), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 92/338 (27%), Positives = 139/338 (41%), Gaps = 9/338 (2%)
Query 14 TQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVLLSNGNKTEEGLVHGSPDRHFAV 73
TQ E RR D P + A + + + A K Y +L SN N EE + G FA
Sbjct 141 TQMEPTSARRAFPCWDEPALKATFTIDITA-KENYTIL-SNMNAVEETVKDGLKTARFA- 197
Query 74 FTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMHSVKVSMK 133
+ +YL A + E + K + + VY+ P + ++A ++
Sbjct 198 --ETCRMSTYLLAWIVAELEYVEYFTPGKHCPRLPVRVYTTPGFSEQGKFAAELGAKTLD 255
Query 134 WEEEHFGREYDLDIFNVACVSDFNAGAMENKGLNIFNCTLLLADMQTSTDDDFERVLAVV 193
+ FG Y L ++ + DF AGAMEN GL + +L + S ERV VV
Sbjct 256 FFSGVFGEPYPLPKCDMVAIPDFEAGAMENWGLVTYRLAAILVS-EDSAATVIERVAEVV 314
Query 194 GHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGTVMSAGVQRIKEVADLISRQFAE 253
HE H W GN VT++ W L L EG + V D + +
Sbjct 315 QHELAHQWFGNLVTMQFWDGLWLNEGFATWMSWFSCNHFYPEWKVWESYVTDNLQSALSL 374
Query 254 DDGPMAHPIR-PESY-MAMDNFYTATVYDKGAEVVRIYHTLLGASGFRKGMDMYFERHDN 311
D +HPI P + ++ + A Y KG+ V+R+ +G F KG+ Y +H
Sbjct 375 DALRSSHPIEVPIMHDYEINQIFDAISYSKGSCVIRMVSKYVGEDTFIKGIQKYISKHRY 434
Query 312 TAATCDDFRAAM-ADANRINLDQMDRWYSQAGTPRLEV 348
+D AA+ A++ + M W + G P L V
Sbjct 435 GNTVTEDLWAALSAESGQDISSTMHNWTKKTGYPVLSV 472
> 7301771
Length=1082
Score = 98.6 bits (244), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 91/348 (26%), Positives = 140/348 (40%), Gaps = 17/348 (4%)
Query 14 TQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVLLSNGNKTEEGLVHGSPDRHFAV 73
TQ E R+ D P A + V L +K LSN E + +
Sbjct 162 TQFEPSSARKAFPCFDEPGFKASFVVTLGYHKQFNA--LSNMPVREIRPHESLANYIWCE 219
Query 74 FTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMHSVKVSMK 133
F + P +YL A +F+ N ++ P + + ++A ++
Sbjct 220 FQESVPMSTYLVAYSVNDFSFKPSTLP----NGALFRTWARPNAIDQCDYAAEFGPKVLQ 275
Query 134 WEEEHFGREYDLDIFNVACVSDFNAGAMENKGLNIFNCTLLLADMQTSTDDDFERVLAVV 193
+ EE FG Y L + V DF+AGAMEN GL + + LL S+ D + + V+
Sbjct 276 YYEEFFGIRYPLPKIDQMAVPDFSAGAMENWGLVKYRESTLLYSPTHSSLADKQDLANVI 335
Query 194 GHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGTVMSAGVQRIKEVADLISRQFAE 253
HE H W GN VT++ W L L EG + L + + R K + F
Sbjct 336 AHELAHQWFGNLVTMKWWTDLWLNEGFATYVAGLGVQEIYPEWHSRDKGSLTALMTAFRL 395
Query 254 DDGPMAHPI-RP-ESYMAMDNFYTATVYDKGAEVVRIYHTLLGASGFRKGMDMYFERHDN 311
D +HPI RP + ++ + A Y KG+ V+R+ H +G FR G+ Y + +
Sbjct 396 DSLVSSHPISRPIQMVTEIEESFDAISYQKGSAVLRMMHLFMGEESFRTGLREYLKLYAY 455
Query 312 TAATCDDFRAAMADANRIN---------LDQMDRWYSQAGTPRLEVLR 350
A ++ ++ A N MD W Q G P L + R
Sbjct 456 KNAEQNNLWESLTTAAHQNGALPGHYDINTIMDSWTLQTGFPVLNITR 503
> Hs7019561
Length=1024
Score = 97.1 bits (240), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 76/270 (28%), Positives = 131/270 (48%), Gaps = 17/270 (6%)
Query 100 TTKSGNEVALAVYSEPAQLHKL--EWAMHSVKVSMKWEEEHFGREYDLDIFNVACVSDFN 157
TTKSG V + +Y+ P + + ++A+H K +++ E++F Y L ++ V
Sbjct 345 TTKSG--VVVRLYARPDAIRRGSGDYALHITKRLIEFYEDYFKVPYSLPKLDLLAVPKHP 402
Query 158 AGAMENKGLNIFNCTLLLADMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTLK 217
AMEN GL+IF +L D S+ V V+ HE H W G+ VT W + LK
Sbjct 403 YAAMENWGLSIFVEQRILLDPSVSSISYLLDVTMVIVHEICHQWFGDLVTPVWWEDVWLK 462
Query 218 EGLTVFREQLFMGT-VMSAG--VQRIKEVADLISRQFAEDDGPMAHPIRPESYMA--MDN 272
EG + E F+GT + G +++ + + D++ D +HP+ E A +D
Sbjct 463 EGFAHYFE--FVGTDYLYPGWNMEKQRFLTDVLHEVMLLDGLASSHPVSQEVLQATDIDR 520
Query 273 FYTATVYDKGAEVVRIYHTLLGASGFRKGMDMYFERHDNTAATCDDFRAAMADANR---- 328
+ Y KGA ++R+ +G S F++G+ Y H A +D +++A +
Sbjct 521 VFDWIAYKKGAALIRMLANFMGHSVFQRGLQDYLTIHKYGNAARNDLWNTLSEALKRNGK 580
Query 329 -INLDQ-MDRWYSQAGTPRLEVLRSEYKED 356
+N+ + MD+W Q G P + +L + E+
Sbjct 581 YVNIQEVMDQWTLQMGYPVITILGNTTAEN 610
> CE27507
Length=1073
Score = 96.3 bits (238), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 94/362 (25%), Positives = 156/362 (43%), Gaps = 27/362 (7%)
Query 9 GSALVTQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVLLSNGNKTEEGLVHGSPD 68
++ T+ E R D P V A + + + NK KY VL SN E H S +
Sbjct 235 ATSFTTKFEPTLARAFFPCWDEPGVKATFNISVRHNK-KYTVL-SNMPPVESH-DHKSWE 291
Query 69 RHF--AVFTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMH 126
F VF P +YL A GEF K +++ + + V++ P + +++ +
Sbjct 292 DQFKTTVFQTTPPMSTYLLAFAIGEFV----KLESRTERGIPVTVWTYPEDVMSMKFTLE 347
Query 127 SVKVSMKWEEEHFGREYDLDIFNVACVSDFNAGAMENKGLNIFNC-----TLLLADMQTS 181
V E+ Y L ++ +F+ G MEN GL +F T + D
Sbjct 348 YAPVIFDRLEDALEIPYPLPKVDLIAARNFHVGGMENWGLVVFEFASIAYTPPITDHVNE 407
Query 182 TDDDFE---RVLAVVGHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGT--VMSAG 236
T D R+ ++ HE H W GN VT+RDW +L L EG F M T ++A
Sbjct 408 TVDRMYNEFRIGKLIAHEAAHQWFGNLVTMRDWSELFLNEGFATFYVYEMMSTERPVTAQ 467
Query 237 VQRIKEVADLISRQFAEDDG-PMAHPIRPESYMAMDNFYTATVYDKGAEVVRIYHTLLGA 295
+ +A LI Q ED + + ES + +F+ +Y KG ++R+ L+
Sbjct 468 FEYYDSLASLILAQSEEDHRLSLVRELATESQVE-SSFHPTNLYTKGCVLIRMLRDLVSD 526
Query 296 SGFRKGMDMYFERHDNTAATCDDFRAAM---AD--ANRINLDQ-MDRWYSQAGTPRLEVL 349
F+ + Y ++ + + DD A++ AD A + L ++ W+ G P + ++
Sbjct 527 FDFKAAVRRYLRKNAYRSVSRDDLFASLPAYADHGAEQEKLSHVLEGWFVNEGIPEITLI 586
Query 350 RS 351
R+
Sbjct 587 RN 588
> Hs11641261
Length=960
Score = 95.1 bits (235), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 64/248 (25%), Positives = 120/248 (48%), Gaps = 9/248 (3%)
Query 82 SYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMHSVKVSMKWEEEHFGR 141
+YL A + +F ++ T SG V +++Y+ P + ++ +A+ + + + E++F
Sbjct 261 TYLVAYIVCDFHSLSG--FTSSG--VKVSIYASPDKRNQTHYALQASLKLLDFYEKYFDI 316
Query 142 EYDLDIFNVACVSDFNAGAMENKGLNIFNCTLLLADMQTSTDDDFERVLAVVGHEYFHNW 201
Y L ++ + DF GAMEN GL + T LL D +TS+ D V V+ HE H W
Sbjct 317 YYPLSKLDLIAIPDFAPGAMENWGLITYRETSLLFDPKTSSASDKLWVTRVIAHELAHQW 376
Query 202 TGNRVTVRDWFQLTLKEGLTVFREQLFMGTV---MSAGVQRIKEVADLISRQFAEDDGPM 258
GN VT+ W + LKEG + E + + + + ++I++ P+
Sbjct 377 FGNLVTMEWWNDIWLKEGFAKYMELIAVNATYPELQFDDYFLNVCFEVITKDSLNSSRPI 436
Query 259 AHPIRPESYMAMDNFYTATVYDKGAEVVRIYHTLLGASGFRKGMDMYFERHDNTAATCDD 318
+ P E+ + + Y+KGA ++ + LG F+KG+ Y ++ A DD
Sbjct 437 SKP--AETPTQIQEMFDEVSYNKGACILNMLKDFLGEEKFQKGIIQYLKKFSYRNAKNDD 494
Query 319 FRAAMADA 326
++++++
Sbjct 495 LWSSLSNS 502
> 7300768
Length=1878
Score = 93.2 bits (230), Expect = 8e-19, Method: Compositional matrix adjust.
Identities = 91/356 (25%), Positives = 139/356 (39%), Gaps = 37/356 (10%)
Query 13 VTQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVLLSNGNKTEEGLVHGSPDRHFA 72
TQ E+ R D P A++ + ++ + + N V S
Sbjct 1112 TTQFESTDARHAFPCYDEPSKRAEFTITIKHDPSY--------NAISNMPVDSSSTSGVT 1163
Query 73 VFTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMHSVKVSM 132
VF PSYL A + EF F+ N + V+S H+ EWA+ + +
Sbjct 1164 VFQKTVNMPSYLVAFIVSEFV-----FSEGELNGLPQRVFSRNGTEHEQEWALTTGMLVE 1218
Query 133 KWEEEHFGREYDLDIFNVACVSDFNAGAMENKGLNIFNCTLLLADMQTSTDDDFERVLAV 192
K +F + L + A + DF AGAMEN GL + LL + + ST + +
Sbjct 1219 KRLSGYFDVPFALPKLDQAGIPDFAAGAMENWGLATYREEYLLYNTENSTTSTQTNIATI 1278
Query 193 VGHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMG---------TVMSAGVQRIKEV 243
HE H W G+ V + W L LKEG E L + AG + V
Sbjct 1279 EAHEDAHMWFGDLVAIEWWSFLWLKEGFATLFENLAVDLAYPEWDIFQTFHAGSYQSALV 1338
Query 244 ADLISRQFAEDDGPMAHPIRPESYMAMDNFYTATVYDKGAEVVRIYHTLLGASGFRKGMD 303
D + + PM+H ++ S +A+ Y + Y K V+ ++ L + F++G+
Sbjct 1339 NDA-----SANARPMSHFVQKPSEIAL--LYDSVSYAKAGSVLDMWRHALTNTVFQRGLH 1391
Query 304 MYFERHDNTAATCDDFRAAMADA--------NRINLDQMDRWYSQAGTPRLEVLRS 351
Y + TAA A+A A D M W +Q G P L V R+
Sbjct 1392 NYLVDNQFTAANPSKLFEAIAKAAVEENYEVKATVPDMMGSWTNQGGVPLLTVTRN 1447
Score = 89.4 bits (220), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 81/288 (28%), Positives = 126/288 (43%), Gaps = 24/288 (8%)
Query 79 PKPS-YLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMHSVKVSMKWEEE 137
PK S YL A + +F + TT N + V+S + + EWA+ S +
Sbjct 246 PKMSTYLVAFIVSDFES-----TTGELNGIRQRVFSRKGKQDQQEWALWSGLLVESSLAS 300
Query 138 HFGREYDLDIFNVACVSDFNAGAMENKGLNIFNCTLLLADMQTSTDDDFERVLAVVGHEY 197
+FG + L + A + DF+AGAMEN GL + + + Q ST + + ++GHEY
Sbjct 301 YFGVPFALPKLDQAGIPDFSAGAMENWGLATYREQYMWWNKQNSTINLKTNIANIIGHEY 360
Query 198 FHNWTGNRVTVRDWFQLTLKEGL-TVFREQLFMGTVMSAGVQRIKEVADLISRQFAE--- 253
H W G+ V+++ W L LKEG T+F + +I V D S +
Sbjct 361 AHMWFGDLVSIKWWTYLWLKEGFATLFSYESNDIAFPLWDTYQIFHVNDYNSALLNDALA 420
Query 254 DDGPMAHPIRPESYMAMDNFYTATVYDKGAEVVRIYHTLLGASGFRKGMDMYFERHDNTA 313
PM H ++ S + N Y Y K A V+ ++ FR G++ Y ++ T+
Sbjct 421 SAVPMTHYVQTPS--EISNRYNTFSYAKPASVLYMFKNAWTDKVFRTGLNKYLTKNQYTS 478
Query 314 ATCDD---FRAAMADANRINL-------DQMDRWYSQAGTPRLEVLRS 351
CD+ F + A+ + D W QAG P L V R+
Sbjct 479 --CDEWDLFASFQESADELGFTLPASVDDIFSSWSHQAGYPLLTVTRN 524
> CE18904
Length=786
Score = 91.3 bits (225), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 88/346 (25%), Positives = 148/346 (42%), Gaps = 25/346 (7%)
Query 13 VTQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVLLSNGNKTEEGLVHGSPDRHFA 72
VTQ E R + D P+ A + V + LSN + + + D
Sbjct 160 VTQFEPTAARFMVPCFDEPEFKAIWHVTVVHPTGS--TALSNAKEIDN--TKTNDDFSTT 215
Query 73 VFTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMHSVKVSM 132
F SY+ AI G+ + TK+G V + VYS+P + ++ A++ ++ +
Sbjct 216 EFESTLKMSSYILAIFVGDVQF--KEAVTKNG--VRIRVYSDPGHIDSVDHALNVSRIVL 271
Query 133 KWEEEHFGREYDLDIFNVACVSDFNAGAMENKGLNIFNCTLLLADMQTSTDDDFERVLAV 192
+ E+ FG Y++D ++ V +F GAMEN GL + L+ ++ D + V
Sbjct 272 EGFEKQFGYPYEMDKLDLIAVYNFRYGAMENWGLIVHQAYTLIENLMPGNTD---IISEV 328
Query 193 VGHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGTVMSAGVQR-IKEVADLISRQF 251
V HE H W GN VT++ W QL L EG + + T + R ++ Q
Sbjct 329 VAHEIAHQWFGNLVTMKFWDQLWLNEGFATYMTA-YGYTFLDPNYDRDYYYLSPQFESQS 387
Query 252 AEDDGPMAHPIRPESYMAMDNFYTATVYDKGAEVVRIYHTLLGASGFRKGMDMYFERHDN 311
+ GP+ I+ S+ A+ N + Y KG +R+ ++G F + + Y HD
Sbjct 388 GNELGPLNSMIKL-SHGAIGN--PSQNYAKGCSFIRMLEKIVGTEHFNEAIRTYLHTHDF 444
Query 312 TAATCDDFRAAM---ADANRIN-----LDQMDR-WYSQAGTPRLEV 348
+ + A+ +D IN +++ R W Q G P + V
Sbjct 445 SNVEDKNLYDALRVYSDKLEINGKPFDIEEFGRCWTHQTGYPTVYV 490
> CE18905
Length=747
Score = 90.1 bits (222), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 89/346 (25%), Positives = 147/346 (42%), Gaps = 34/346 (9%)
Query 14 TQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVLLSNGNKTEEGLVHGSPDRHFAV 73
TQ E R + D P+ A + V LE LSNG + E + + D
Sbjct 164 TQFETIFARNMIPCFDEPEFKATWNVSLEHPTGS--TALSNGIEVESKV---NDDWKTTT 218
Query 74 FTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMHSVKVSMK 133
+ SY+ A+ G+ I+ K T + N V + VY++P + +++ A++ ++ ++
Sbjct 219 YKKTLKMSSYILALFIGD---IQFKETILN-NGVRIRVYTDPVNIDRVDHALNISRIVLE 274
Query 134 WEEEHFGREYDLDIFNVACVSDFNAGAMENKGLNIFNCTLLLADMQTSTDDDFERVLAVV 193
E FG Y ++ + V +F GAMEN GL I N L+ D T+ +V
Sbjct 275 GFERQFGIRYPMEKLDFVSVQNFKFGAMENWGLVIHNAYSLIGDPMDVTE--------IV 326
Query 194 GHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGTVMSAGVQRIKEVADLISRQFAE 253
HE H W GN VT++ W + L EG + + T + QR ++ QF
Sbjct 327 IHEIAHQWFGNLVTMKYWDHIWLNEGFASYMTSYGL-TFLDPDYQR---DTFYLNCQFNP 382
Query 254 --DDGPMAHPIRPESYMAMDNFYTATVYDKGAEVVRIYHTLLGASGFRKGMDMYFERHDN 311
+DG ++ M+N ATVY KG VR+ + G F + + Y E++
Sbjct 383 QINDGGVSLNFGKNLSSVMNN--AATVYMKGCSFVRMLENIFGTEYFNEAIKFYLEKNQY 440
Query 312 TAATCDDFRAAM--------ADANRINLDQMDR-WYSQAGTPRLEV 348
+D A + ++++ R W Q G P ++V
Sbjct 441 DNVDDNDLYEAFRTSSETLDTNGKPFDIEKFARCWTHQNGFPTIQV 486
> 7300769
Length=1646
Score = 88.2 bits (217), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 82/314 (26%), Positives = 136/314 (43%), Gaps = 40/314 (12%)
Query 14 TQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPV----LLSNGNKTEEGLVHGSPDR 69
TQ + RR D P A + V L+ ++ V L+S+ TEEG+
Sbjct 180 TQFQTHHARRAFPSFDEPQFKATFDVTLKRHRTFNSVSNTRLISSYPSTEEGI------- 232
Query 70 HFAVFTDPHPKPS-YLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMHSV 128
F+ PK S YL A + EF A +D VY+ P + ++ ++V
Sbjct 233 -FSDVYKTTPKMSTYLLAFIISEFVARKDD---------DFGVYARPEYYAQTQYP-YNV 281
Query 129 KVSMKWEEEHFGREYDLDIF-------NVACVSDFNAGAMENKGLNIFNCTLLLADMQTS 181
+ + E G+ D D + ++A + DF+AGAMEN GL + LL D +
Sbjct 282 GIQIL---EEMGQYLDKDYYSMGNDKMDMAAIPDFSAGAMENWGLLTYRERSLLVDESAT 338
Query 182 TDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGTVMSAGVQRIK 241
T + + AVV HE H W G+ VT + W L EG + + + GT M ++
Sbjct 339 TLASRQSIAAVVAHEQAHMWFGDLVTCKWWSYTWLNEGFARYFQ--YFGTAMVEDKWELE 396
Query 242 E--VADLISRQFAEDDGPMAHPIRPESYMA---MDNFYTATVYDKGAEVVRIYHTLLGAS 296
+ V D + A D +P+ E+ + + + Y+KGA +R+ +G
Sbjct 397 KQFVVDQVQSVMAMDSTNATNPLSDENTYTPAHLSRMFNSISYNKGATFIRMIKHTMGEQ 456
Query 297 GFRKGMDMYFERHD 310
F+K + Y ++++
Sbjct 457 QFQKSLQEYLKKYE 470
Score = 67.4 bits (163), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 72/309 (23%), Positives = 124/309 (40%), Gaps = 27/309 (8%)
Query 13 VTQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVLLSNGNKTEEGLVHGSPDRHFA 72
+TQ + R + D P + A++++++ Y + + K + L S DR
Sbjct 1112 LTQMQRINARLVLPCFDEPALKAQFQLQI-VRPNGYQSIANTKLKETKAL---SQDRFVD 1167
Query 73 VFTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMHSVKVSM 132
F + +YL A + ++A GNE AV + P E++ H + +
Sbjct 1168 HFKETPVMSTYLLAFMVANYSA--------RGNESEFAVLTRPEFYDNTEFSYHVGQQVV 1219
Query 133 KWEEEHFGREY---DLDIFNVACVSDFNAGAMENKGLNIFNCTLLLADMQTSTD-DDFER 188
E F Y D+ A F MEN GL I++ +L+ + S D D E
Sbjct 1220 SAYGELFQSPYAELGNDVLQYASSPRFPHNGMENWGLIIYSDDVLIQEPGYSDDWSDKEF 1279
Query 189 VLAVVGHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGTVMSAGVQR-----IKEV 243
+ ++ HE H W G+ VT W L E + E FM + ++++
Sbjct 1280 AIRIIAHETSHMWFGDSVTFSWWSYFWLNEAFARYYE-YFMAHQLYPDYHLDEQFVVRQM 1338
Query 244 ADLISRQFAEDDGPMAHP---IRPESYMAMDNFYTATVYDKGAEVVRIYHTLLGASGFRK 300
+ PM P I+ S +A ++ Y KGA +VR++ L+GA F
Sbjct 1339 QLIFGTDAQNGTQPMTSPESEIQTPSQIAYK--FSGIAYAKGACIVRMWRNLMGAENFDT 1396
Query 301 GMDMYFERH 309
+ Y +++
Sbjct 1397 AIRSYLQQY 1405
> CE26001
Length=1815
Score = 84.7 bits (208), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 81/327 (24%), Positives = 140/327 (42%), Gaps = 26/327 (7%)
Query 12 LVTQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVLLSNGNKTEEGLVHGSPDRHF 71
+ T E R++ LD P AK+ + ++ ++ V LSN +TE + D +
Sbjct 215 IATHMEPFSARKVFPSLDEPSYKAKFTITVQYPASQ--VALSNMMETEPTKI----DNIW 268
Query 72 AVFTDPH-PK-PSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMHSVK 129
+ T P PK SYL A G + + ++ K + + P L++A +
Sbjct 269 STITFPQTPKMSSYLIAFAVGPY--VNSQYVNK--HNTLTRAWGWPGTEQYLQFAAQNAG 324
Query 130 VSMKWEEEHFGREYDLDIFNVACVSDFNAGAMENKGLNIFNCTLLLADMQTSTDDDFERV 189
+ E+ G ++ L + + +F AGAMEN GL I+ + + T T + E
Sbjct 325 ECLYQLGEYTGIKFPLSKADQLGMPEFLAGAMENWGLIIYKYQYIAYNPTTMTTRNMEAA 384
Query 190 LAVVGHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGTVMSA------GVQRIKEV 243
V+ HE H W G+ VT W L L EG + +V +Q + E+
Sbjct 385 AKVMCHELAHQWFGDLVTTAWWDDLFLNEGFADYFMTFIQKSVYPQQATYLDTLQVLDEL 444
Query 244 ADLISRQFAEDDGPMAHPIRPESYMAMDNFYTATVYDKGAEVVRIYHTLLGASGFRKGMD 303
++ D P+ +P P A D+ Y+KGA ++R+ +LGA F++G+
Sbjct 445 QVGLTADVRYDAHPLVYPDGP----AFDDI----TYNKGASMLRMLSDVLGADVFKQGIR 496
Query 304 MYFERHDNTAATCDDFRAAMADANRIN 330
Y ++ + A D + + D + N
Sbjct 497 AYLQKMQYSNANDFDLFSTLTDTAKSN 523
Score = 60.8 bits (146), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 41/141 (29%), Positives = 68/141 (48%), Gaps = 6/141 (4%)
Query 82 SYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMHSVKVSMKWEEEHFGR 141
+YL A+ G F+ + T++G V V++ E+A++ ++ + E +F
Sbjct 1205 TYLLALCVGHFSNLAT--VTRTG--VLTRVWTWSGMEQYGEFALNVTAGTIDFMENYFSY 1260
Query 142 EYDLDIFNVACVSDF--NAGAMENKGLNIFNCTLLLADMQTSTDDDFERVLAVVGHEYFH 199
++ L +V + ++ NAGAMEN GL I +L + D +T D V HE H
Sbjct 1261 DFPLKKLDVMALPEYTMNAGAMENWGLIIGEYSLFMFDPDYATTRDITEVAETTAHEVVH 1320
Query 200 NWTGNRVTVRDWFQLTLKEGL 220
W G+ VT+ W + L EG
Sbjct 1321 QWFGDIVTLDWWNDIFLNEGF 1341
> SPCC1322.05c
Length=612
Score = 83.2 bits (204), Expect = 8e-16, Method: Compositional matrix adjust.
Identities = 83/353 (23%), Positives = 144/353 (40%), Gaps = 44/353 (12%)
Query 12 LVTQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVLLSNGNKTEEGLVHGSPDRHF 71
+ ++C+A R D P V ++ ++K PV+ S +GS +
Sbjct 133 VFSECQAIHARSFIPCQDTPSVKVPCTFKI---RSKLPVIASGIPCGTANFCNGSLEY-- 187
Query 72 AVFTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAM-HSVKV 130
+F +P PSYLF IL+G+ A +T G + VY+EP L ++ H ++
Sbjct 188 -LFEQKNPIPSYLFCILSGDLA------STNIGPRSS--VYTEPGNLLACKYEFEHDMEN 238
Query 131 SMKWEEE----HFGREYDLDIFNVACVSDFNAGAMENKGLNIFNCTLLLADMQTSTDDDF 186
M+ E+ + YD I + F G MEN TL+ D
Sbjct 239 FMEAAEQLTLPYCWTRYDFVILPPS----FPYGGMENPNATFATPTLIAGDRSN------ 288
Query 187 ERVLAVVGHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGTVMSAGVQRIKEVADL 246
+ V+ HE H+W+GN VT W L EG+TVF E+ +G + ++ + +
Sbjct 289 ---VNVIAHELAHSWSGNLVTNESWQCFWLNEGMTVFLERKILGRLYGEPTRQFEAIIGW 345
Query 247 ----ISRQFAEDDGPMAHPIRPESYMAMDNFYTATVYDKGAEVV-RIYHTLLGASGFRKG 301
S + +D I+ D+ ++ Y+KG+ + I + G S F
Sbjct 346 GELEESVKLLGEDSEYTKLIQNLEGRDPDDAFSTVPYEKGSNFLYEIERVIGGPSVFEPF 405
Query 302 MDMYFERHDNTAATCDDFRAAMAD-------ANRINLDQMDRWYSQAGTPRLE 347
+ YF + + F+ A+ + A++++ D WY G P ++
Sbjct 406 LPFYFRKFAKSTVNEVKFKHALYEYFSPLGLASKLDSIDWDTWYHAPGMPPVK 458
> Hs4505029
Length=611
Score = 80.5 bits (197), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 87/353 (24%), Positives = 137/353 (38%), Gaps = 34/353 (9%)
Query 12 LVTQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVLLSNGNKTEEGLVHGSPDRHF 71
L +QC+A R I D P V Y E + K V L + + E P R
Sbjct 132 LFSQCQAIHCRAILPCQDTPSVKLTYTA--EVSVPKELVALMSAIRDGETPDPEDPSRKI 189
Query 72 AVFTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMHSVKVS 131
F P P YL A++ G + + T V+SE Q+ K + +
Sbjct 190 YKFIQKVPIPCYLIALVVGALESRQIGPRT--------LVWSEKEQVEKSAYEFSETESM 241
Query 132 MKWEEEHFGR----EYDLDIFNVACVSDFNAGAMENKGLNIFNCTLLLADMQTSTDDDFE 187
+K E+ G +YDL + + F G MEN L TLL D S
Sbjct 242 LKIAEDLGGPYVWGQYDLLVLPPS----FPYGGMENPCLTFVTPTLLAGDKSLSN----- 292
Query 188 RVLAVVGHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGTVMSAGVQRIKEVADLI 247
V+ HE H+WTGN VT + W L EG TV+ E+ G + + +
Sbjct 293 ----VIAHEISHSWTGNLVTNKTWDHFWLNEGHTVYLERHICGRLFGEKFRHFNALGGWG 348
Query 248 SRQFAEDDGPMAHP----IRPESYMAMDNFYTATVYDKGAEVVRIYHTLLGASGFRKG-M 302
Q + HP + + + D Y++ Y+KG ++ LLG G +
Sbjct 349 ELQNSVKTFGETHPFTKLVVDLTDIDPDVAYSSVPYEKGFALLFYLEQLLGGPEIFLGFL 408
Query 303 DMYFERHDNTAATCDDFRAAMADANRINLDQMDR--WYSQAGTPRLEVLRSEY 353
Y E+ + T DD++ + + +D +++ W + +P L ++ Y
Sbjct 409 KAYVEKFSYKSITTDDWKDFLYSYFKDKVDVLNQVDWNAWLYSPGLPPIKPNY 461
> CE24087
Length=781
Score = 80.1 bits (196), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 84/372 (22%), Positives = 163/372 (43%), Gaps = 43/372 (11%)
Query 3 KGLYVSGSALVTQCEAEGFRRITYFLDRPDVLAKYKVRLEAN-----KAKYPVLLSNGNK 57
+ LY SA TQ E R++ D P+ A ++V + N ++ +L+S K
Sbjct 11 ENLYFRKSA-ATQFEPTFARKMLPCFDEPNFKATFQVAIIRNPHHIARSNMNILISKEYK 69
Query 58 TEEGLVHGSPDRHFAVFTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQ 117
GL+ VF +YL A+ + + T + + + +Y+
Sbjct 70 N--GLIKD-------VFEKSVKMSTYLLAVAVLDGYGYIKRLTRNTQKAIEVRLYAPQDM 120
Query 118 LH-KLEWAMHSVKVSMKWEEEHFGREYDLDIFNVACVSDFNAGAMENKGLNIFNCTLLLA 176
L + E+ + + ++++ E++F Y LD ++ + DF+ GAMEN GL F + LL
Sbjct 121 LTGQSEFGLDTTIRALEFFEDYFNISYPLDKIDLLALDDFSEGAMENWGLVTFRDSALLF 180
Query 177 DMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGTVMSAG 236
+ + ++ E + ++ HE H W GN VT+ W ++ L EG + E
Sbjct 181 NERKASVVAKEHIALIICHEIAHQWFGNLVTMDWWNEVFLNEGFANYMEY--------KC 232
Query 237 VQRIKEVADLISRQFAEDDGPMAHPIRPESYM-----------AMDNFYTATVYDKGAEV 285
V+ + ++SR +AE+ +A P+ ++ ++ N + A Y K A +
Sbjct 233 VEHLFPDWSIMSRFYAEN---LAFSQEPDGFLSSRAIESDDDDSLLNLFDAINYHKAAAI 289
Query 286 VRIYHTLLGASGFRKGMDMYFERHD-NTAATCDDFRAAMADAN---RINLDQMDRWY-SQ 340
+ + + G F+ + Y ++ + A D ++ AN +++ + + Y +Q
Sbjct 290 IHMIAEMAGQKNFQNALVEYLNKYAYSNAIGVDLWKIVEKHANLQGTVSIPDLAKAYTTQ 349
Query 341 AGTPRLEVLRSE 352
G P L V R++
Sbjct 350 VGYPLLTVERTD 361
> CE24840
Length=609
Score = 80.1 bits (196), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 87/360 (24%), Positives = 144/360 (40%), Gaps = 62/360 (17%)
Query 12 LVTQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVLLSNGNKTEEGLVHGSPDRHF 71
L +QC+A R I +D P V + Y+ + ++ + G + R
Sbjct 134 LFSQCQAINARSIVPCMDTPSVKSTYEAEVCVPIGLTCLMSAIGQGSTPSECG---KRTI 190
Query 72 AVFTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMHSVKVS 131
F P PSYL AI+ G R + + + AV++EP+Q + +
Sbjct 191 FSFKQPVSIPSYLLAIVVGHLE--RKEISERC------AVWAEPSQAEASFYEFAETEKI 242
Query 132 MKWEEEHFGR----EYDLDIFNVACVSDFNAGAMENKGLNIFNCTLLLADMQTSTDDDFE 187
+K E+ G YDL V + F G MEN L TLL D
Sbjct 243 LKVAEDVAGPYVWGRYDL----VVLPATFPFGGMENPCLTFITPTLLAGD---------R 289
Query 188 RVLAVVGHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGTVMSAGVQRIKEVADLI 247
++ V+ HE H+WTGN VT W L EG TVF E+ G + +
Sbjct 290 SLVNVIAHEISHSWTGNLVTNFSWEHFWLNEGFTVFLERKIHGKMYGE-----------L 338
Query 248 SRQFAEDDGPMAHPIR-------PE-SYMAM---------DNFYTATVYDKGAEVVRIYH 290
RQF + G +R P+ Y + D+ +++ Y+KG+ ++
Sbjct 339 ERQFESESGYEEALVRTVNDVFGPDHEYTKLVQNLGNADPDDAFSSVPYEKGSALLFTIE 398
Query 291 TLLG-ASGFRKGMDMYFERHDNTAATCDDFRAAMADA---NRINLDQMD--RWYSQAGTP 344
LG S F + + Y +++ + ++++ + D+ ++ LD +D W +AG P
Sbjct 399 QALGDNSRFEQFLRDYIQKYAYKTVSTEEWKEYLYDSFTDKKVILDNIDWNLWLHKAGLP 458
> CE28160
Length=625
Score = 79.3 bits (194), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 62/226 (27%), Positives = 98/226 (43%), Gaps = 28/226 (12%)
Query 12 LVTQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVLLSNGNKTEEGLVHGSPDRHF 71
L +QC+A R I +D P V + Y+ + ++ + G ++ G D
Sbjct 138 LFSQCQAIHARSIVPCMDTPSVKSTYEAEVTVPTGMTCLMSAIGQGSK-----GDDDTTT 192
Query 72 AVFTDPHPKPSYLFAILAG--EFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMHSVK 129
+ P PSYL AI+ G E I D+ AV++EP+ + K W +
Sbjct 193 FFYKQPVAIPSYLIAIVVGCLEKRDISDR----------CAVWAEPSVVDKAHWEFAETE 242
Query 130 VSMKWEEEHFGREYDLDIFNVACVS-DFNAGAMENKGLNIFNCTLLLADMQTSTDDDFER 188
+ EE G+ Y +++ C+ F G MEN L TL+ D
Sbjct 243 DILASAEEIAGK-YIWGRYDMVCLPPSFPFGGMENPCLTFLTPTLIAGD---------RS 292
Query 189 VLAVVGHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGTVMS 234
+++V+ HE H+W+GN VT W L EG T+F E+ G ++S
Sbjct 293 LVSVIAHEIAHSWSGNLVTNSSWEHFWLNEGFTMFIERKICGRLVS 338
> 7301517
Length=1078
Score = 79.3 bits (194), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 85/362 (23%), Positives = 144/362 (39%), Gaps = 36/362 (9%)
Query 12 LVTQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVLLSNGNKT-----EEGLVHGS 66
+ TQ RR DRPD+ A + + + ++ + LSN K+ G +
Sbjct 311 ISTQFSPVDARRAFPCFDRPDMKANFSISI-VRPMQFKMALSNMPKSGSRRFRRGFIRDD 369
Query 67 PDRHFAVFTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMH 126
F P+YL A + R + SG + +++ P + +A
Sbjct 370 -------FETTPKMPTYLVAFIVSNMVDSRLA-SQDSGLTPRVEIWTRPQFVGMTHYAYK 421
Query 127 SVKVSMKWEEEHFGREYDLDIFNVACVSDFNAGAMENKGLNIFNCTLLLA--DMQTSTDD 184
V+ + + E+ FG + L ++ V DF AMEN GL F + LL D+Q ++
Sbjct 422 MVRKFLPYYEDFFGIKNKLPKIDLVSVPDFGFAAMENWGLITFRDSALLVPEDLQLASSS 481
Query 185 DFERVLA-VVGHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGTVMSAGVQRIKEV 243
+ +V+A ++ HE H W GN VT + W L LKEG + + + +
Sbjct 482 EHMQVVAGIIAHELAHQWFGNLVTPKWWDDLWLKEGFACYMSY----KALEHAHPEFQSM 537
Query 244 ADLISRQFAE----DDGPMAHPIRPESYMAMD--NFYTATVYDKGAEVVRIYHTLLGASG 297
L +F E D +H I + D + Y KG ++R+ ++++G
Sbjct 538 DTLTMLEFKESMEHDADNTSHAISFDVRSTNDVRRIFDPISYSKGTILLRMLNSIVGDVA 597
Query 298 FRKGMDMYFERHDNTAATCDDFRAAMAD--------ANRINLDQ-MDRWYSQAGTPRLEV 348
FR ++ DD A + +++ Q MD W +Q G P + V
Sbjct 598 FRSATRDLLKKFAYGNMDRDDLWAMLTRHGHEQGTLPKDLSVKQIMDSWITQPGYPVVNV 657
Query 349 LR 350
R
Sbjct 658 ER 659
> At4g33090
Length=873
Score = 76.3 bits (186), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 90/337 (26%), Positives = 128/337 (37%), Gaps = 51/337 (15%)
Query 13 VTQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVLLSNGNKTEEGLVHGSPDRHFA 72
VTQ E RR D P A +K+ LE V LSN EE V+G +
Sbjct 149 VTQFEPADARRCFPCWDEPACKATFKITLEVPTDL--VALSNMPIMEEK-VNG--NLKIV 203
Query 73 VFTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMHSVKVSM 132
+ + +YL AI+ G F + D T G + L + S+ S
Sbjct 204 SYQESPIMSTYLVAIVVGLFDYVEDH--TSDGPSLTFETLCACIFLSFFNGCIISLHKS- 260
Query 133 KWEEEHFGREYDLDIFNVACVSDFNAGAMENKGLNIFNCTLLLADMQTSTDDDFERVLAV 192
+F Y L ++ + DF AGAMEN GL + T LL D Q S + +RV +
Sbjct 261 NHSCRYFAVPYPLPKMDMIAIPDFAAGAMENYGLVTYRETALLYDEQHSAASNKQRVSYL 320
Query 193 VGHEYFHNWTGNRVTVRDWFQLT--LKEGLTVFREQLFMGTVMSAGVQRIKEVADLISRQ 250
F W + W Q EGL +
Sbjct 321 ATDSLFPEW-------KIWTQFLDESTEGLRL---------------------------- 345
Query 251 FAEDDGPMAHPIRPESYMA--MDNFYTATVYDKGAEVVRIYHTLLGASGFRKGMDMYFER 308
D +HPI E A +D + A Y KGA V+R+ + LGA F+K + Y +
Sbjct 346 ---DGLEESHPIEVEVNHAAEIDEIFDAISYRKGASVIRMLQSYLGAEVFQKSLAAYIKN 402
Query 309 HDNTAATCDDFRAAMADANRINLDQ-MDRWYSQAGTP 344
H + A +D AA+ + +++ M W Q G P
Sbjct 403 HAYSNAKTEDLWAALEAGSGEPVNKLMSSWTKQKGYP 439
> CE20255
Length=1045
Score = 72.8 bits (177), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 92/371 (24%), Positives = 140/371 (37%), Gaps = 51/371 (13%)
Query 11 ALVTQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPV--LLSNGNKTE----EGLVH 64
+ TQ + R + +D PD+ A++ + + ++ N K + H
Sbjct 283 VVATQLQISEARTVFPCIDVPDMKAQFDTVIIHPTGTTSIANMMENSTKVDGEWTTTTFH 342
Query 65 GSPDRHFAVFTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWA 124
+P P +YLFA ++ + T SG V VY +P +L +
Sbjct 343 RTP-----------PMSTYLFAFSVSDYPYLE----TFSGRGVRSRVYCDPTKLVDAQLI 387
Query 125 MHSVKVSMKWEEEHFGREYDLDIFNVACVSDFNAGAMENKGLNIFNCTLLLADMQTSTDD 184
S+ + + E++FG Y L+ +V V + AMEN GL T L
Sbjct 388 TKSIGPVLDFYEDYFGIPYPLEKLDVVIVPALSVTAMENWGLITIRQTNGLYTEGRFAIS 447
Query 185 DFERVLAVVGHEYFHNWTGNRVTVRDWFQLTLKEG---LTVFREQLFMGTVM------SA 235
V +V HE H W GN VT++ W L L EG L R F+ SA
Sbjct 448 QKHDVQEIVAHELAHQWFGNLVTMKWWNDLWLNEGFATLISVRAVDFLENTTWRYEDSSA 507
Query 236 GVQRIKEVADLISRQFAEDDGPMAHPIRP--ESYMAMDNFYTATVYDKGAEVVRIYHTLL 293
Q + AD I + P++ +SYM + A +Y K A ++R+ L+
Sbjct 508 ESQCVALRADQI-----DSISPVSGKKNSNFDSYMEIQG--KAIIYKKSAIIIRMIERLV 560
Query 294 GASGFRKGMDMYF----------ERHDNTAATCDDFRAA--MADANRINLDQMDRWYSQA 341
FR+G+ + E N D A + N D MD W QA
Sbjct 561 EEDIFRQGLKRFLNTFLYKNADHEDLFNVLVYVHDSSAGGHLRGQNFSLSDVMDTWIRQA 620
Query 342 GTPRLEVLRSE 352
P + V R E
Sbjct 621 HFPVIHVNRKE 631
> 7298498
Length=613
Score = 72.4 bits (176), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 82/350 (23%), Positives = 136/350 (38%), Gaps = 47/350 (13%)
Query 12 LVTQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVLLSNGNKTEEGLVHGSPDRHF 71
+ +QC+A R + D P V Y +E ++ + +K E G
Sbjct 136 MFSQCQAIHARSVIPCQDTPAVKFTYDATVEHPSELTALMSALIDKKEPGK--------- 186
Query 72 AVFTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMHSVKVS 131
+F P P+YL AI G K ++ E + +V++E A +
Sbjct 187 TLFKQEVPIPAYLVAIAIG-------KLVSRPLGENS-SVWAEEAIVDACAEEFSETATM 238
Query 132 MKWEEEHFG----REYDLDIFNVACVSDFNAGAMENKGLNIFNCTLLLADMQTSTDDDFE 187
+K E G ++YDL + F G MEN L TLL D +
Sbjct 239 LKTATELCGPYVWKQYDL----LVMPPSFPFGGMENPCLTFVTPTLLAGD---------K 285
Query 188 RVLAVVGHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGTVMSAGVQRIKEVADLI 247
+ VV HE H+WTGN VT +++ L EG TVF E +G + A K +++L
Sbjct 286 SLADVVAHEIAHSWTGNLVTNKNFEHFWLNEGFTVFVESKIVGRMQGAKELDFKMLSNLT 345
Query 248 SRQFA-----EDDGPMAHPIRPESYMAMDNFYTATVYDKGAEVVRIYHTLLGA-SGFRKG 301
Q + + S D+ +++ Y KG+ +R L G + F
Sbjct 346 DLQECIRTQLNKTPELTKLVVDLSNCGPDDAFSSVPYIKGSTFLRYLEDLFGGPTVFEPF 405
Query 302 MDMYFERHDNTAATCDDFRAAMAD-------ANRINLDQMDRWYSQAGTP 344
+ Y +++ + DF++A+ D ++++ D W G P
Sbjct 406 LRDYLKKYAYKSIETKDFQSALYDYFIDTDKKDKLSAVDWDLWLKSEGMP 455
> 7291558
Length=844
Score = 70.9 bits (172), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 69/278 (24%), Positives = 117/278 (42%), Gaps = 14/278 (5%)
Query 85 FAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMHSVKVSMKWEEEHFGREYD 144
FA+ E AA + +K+ N V + +P L + +M + + E F +
Sbjct 161 FAVHDLENAATEE---SKTSNRVIFRNWMQPKLLGQEMISMEIAPKLLSFYENLFQINFP 217
Query 145 LDIFNVACVSDFNAGAMENKGLNIFNCTLLLADMQTSTDDDFERVLAVVGHEYFHNWTGN 204
L + V AMEN GL +N L + + V HEY H W GN
Sbjct 218 LAKVDQLTVPTHRFTAMENWGLVTYNEERLPQNQGDYPQKQKDSTAFTVAHEYAHQWFGN 277
Query 205 RVTVRDWFQLTLKEGLTVFREQLFMGTVMSAGVQRIKEVADLISRQFAEDDGPMAHPIRP 264
VT+ W L LKEG + + L + ++ + + ++ ++ F++D I
Sbjct 278 LVTMNWWNDLWLKEGPSTYFGYLALDSLQPEWRRGERFISRDLANFFSKDSNATVPAISK 337
Query 265 ESYMAMDNF--YTATVYDKGAEVVRIYHTLLGASGFRKGMDMYFERHD-NTAATCDDFRA 321
+ + +T VY+KG+ +R+ H L+G F G+ + ER A D + +
Sbjct 338 DVKNPAEVLGQFTEYVYEKGSLTIRMLHKLVGEEAFFHGIRSFLERFSFGNVAQADLWNS 397
Query 322 AMADA--NRI-----NLDQ-MDRWYSQAGTPRLEVLRS 351
A N++ NL + MD W Q G P + ++R+
Sbjct 398 LQMAALKNQVISSDFNLSRAMDSWTLQGGYPLVTLIRN 435
> 7291179
Length=931
Score = 69.7 bits (169), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 86/365 (23%), Positives = 141/365 (38%), Gaps = 47/365 (12%)
Query 14 TQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVL---LSNGNKTEEGLVHGS-PDR 69
TQ E R D P +K+ L +P L LSN + + H S D
Sbjct 156 TQFEPNHAREAFPCFDDPIFRTPFKINLA-----HPYLYRALSNM-PVQRTIRHASLKDY 209
Query 70 HFAVFTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMHSVK 129
+ F + HP +YL A + +F R FT+ ++ ++ ++ P L + E+A V
Sbjct 210 VWTQFVESHPMQTYLVAFMISKFD--RPGFTSSERSDCPISTWARPDALSQTEFANMVVA 267
Query 130 VSMKWEEEHFGREYDLDIFNVACVSDFNAGAMENKGLNIFNCTLLLADMQTSTDDDFERV 189
+ + E+ F Y ++ + DF + EN GL F LL D Q S+ DD + V
Sbjct 268 PLLSFYEDLFNSTYRQKKIDLVALPDFTFKSKENWGLPAFAEESLLYDSQRSSIDDQQGV 327
Query 190 LAVVGHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGTVMSAGVQRIKEVADLISR 249
V + W GN V+V W ++ LK ++ + GV ++ D R
Sbjct 328 ARAVAMMVVNQWFGNLVSVAWWHEIWLKNVFALYLSRF--------GVHSLRPEWDYQER 379
Query 250 QFAEDDGPMAHPIRPESYMAMDNFYTATVYD--------------KGAEVVRIYHTLLGA 295
+ + + + TA+V D K A + + H ++G
Sbjct 380 HALQ----LYLSVLDYDAHVNTDLVTASVPDESHIWAAYNEIGERKAAVLFEMLHRVMGE 435
Query 296 SGFRKGMDMYFERHDNTAATCDDFRAAM---ADAN-----RINLDQ-MDRWYSQAGTPRL 346
+ + Y + N AT DF + D N +N+ + M W Q G P +
Sbjct 436 EAWLTALRRYLVVYANRTATSSDFWDLLQLQVDRNGRLGKGLNITRIMKCWLGQPGYPLV 495
Query 347 EVLRS 351
V R+
Sbjct 496 TVTRN 500
> 7300767
Length=988
Score = 68.6 bits (166), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 67/312 (21%), Positives = 127/312 (40%), Gaps = 31/312 (9%)
Query 14 TQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVLLSNGNKTEEGLVHGSPDRHFAV 73
TQCE R I D P + + +++ + + + +E L HG D
Sbjct 164 TQCEPTYGRLIFPCYDEPGFKSNFSIKITHGSSHSAI---SNMPVKEVLAHG--DLKTTS 218
Query 74 FTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMHSVKVSMK 133
F P +YL A + +F +I + + + ++Y+ P K + A+ + ++
Sbjct 219 FHTTPPISTYLVAFVISDFGSISETY-----RGITQSIYTSPTSKEKGQVALKNAVRTVA 273
Query 134 WEEEHFGREYDLDIFNVACVSDFNAGAMENKGLNIFNCTLLLADMQTSTDDDFERVLAVV 193
E++FG Y L + + AMEN GL + LL ++ + D +R L ++
Sbjct 274 ALEDYFGVSYPLPKLDHVALKKNYGAAMENWGLITYKDVNLLKNI---SSDGQKRKLDLI 330
Query 194 --GHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGTVMSAGVQRIKEVADLISRQF 251
HE H W GN V+ W + EG + V++ + ++ D+
Sbjct 331 TQNHEIAHQWFGNLVSPEWWTYTWMNEGFAT-----YFSYVITDLIYPNDKMMDMFMTHE 385
Query 252 AE---------DDGPMAHPIRPESYMAMDNFYTATVYDKGAEVVRIYHTLLGASGFRKGM 302
A+ D PM+H + E + + Y +GA V++++H F +G+
Sbjct 386 ADSAYSYNSFFDVHPMSHYVEGEKDIM--GVFDIISYKRGACVIKMFHHAFRQKLFVRGI 443
Query 303 DMYFERHDNTAA 314
+ E++ + A
Sbjct 444 SHFLEKYRYSVA 455
> YNL045w
Length=671
Score = 67.8 bits (164), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 73/308 (23%), Positives = 123/308 (39%), Gaps = 40/308 (12%)
Query 12 LVTQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVLLSNGNKTEEGLVHGSPDRHF 71
+ +Q EA R + D P V + + +E+ PV+ S G + E+ S D +
Sbjct 181 VFSQLEAIHARSLFPCFDTPSVKSTFTASIES---PLPVVFS-GIRIEDT----SKDTNI 232
Query 72 AVFTDPHPKPSYLFAILAGEF--AAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMHSVK 129
F P P+YL I +G+ A I + T VY+EP +L +W +
Sbjct 233 YRFEQKVPIPAYLIGIASGDLSSAPIGPRST----------VYTEPFRLKDCQWEFENDV 282
Query 130 VSMKWEEEHFGREYDLDIFNVAC-VSDFNAGAMENKGLNIFNCTLLLADMQTSTDDDFER 188
E EY+ +++ V + G ME+ + TL+ D
Sbjct 283 EKFIQTAEKIIFEYEWGTYDILVNVDSYPYGGMESPNMTFATPTLIAHDRSN-------- 334
Query 189 VLAVVGHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGTVMSAGVQRIKEVADLIS 248
+ V+ HE H+W+GN VT W L EG TV+ E+ +G + + +
Sbjct 335 -IDVIAHELAHSWSGNLVTNCSWNHFWLNEGWTVYLERRIIGAIHGEPTRHFSALIGWSD 393
Query 249 RQFAEDDGPMAHPIRPESYMA-------MDNFYTATVYDKGAEVVRIYHTLLGASG-FRK 300
Q + D M P R + + D+ ++ Y+KG ++ T+LG F
Sbjct 394 LQNSIDS--MKDPERFSTLVQNLNDNTDPDDAFSTVPYEKGFNLLFHLETILGGKAEFDP 451
Query 301 GMDMYFER 308
+ YF++
Sbjct 452 FIRHYFKK 459
> At5g13520
Length=616
Score = 53.9 bits (128), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 74/318 (23%), Positives = 121/318 (38%), Gaps = 45/318 (14%)
Query 14 TQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVLLS-----------NGNKTEEGL 62
TQC+A R I D P +Y V + + V+ + E G
Sbjct 127 TQCQAIHARSIFPCQDTPAARIRYDVVMNIPNSLSAVMSARHVRRRLAVPEEAKHLEAGS 186
Query 63 VHGS----PDRHFAVFTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQL 118
+ S DR F P P YLFA GE T+ VY+E A +
Sbjct 187 LGSSLWCGEDRVVEEFAMEQPIPPYLFAFAVGELGFREVGPRTR--------VYTESAAI 238
Query 119 HKLEWA---MHSVKVSMKWEEEHFGREYDLDIFNVACVS-DFNAGAMENKGLNIFNCTLL 174
L+ A + +K E+ FG +Y+ + F++ + F G MEN + T++
Sbjct 239 EVLDAAALEFAGTEDMIKQGEKLFG-DYEWERFDLLVLPPSFPYGGMENPRMVFLTPTVI 297
Query 175 LADMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGTVMS 234
D + VV HE H+WTGN +T + L EG T + E+ + V
Sbjct 298 KGDATGAQ---------VVAHELAHSWTGNLITNINNEHFWLNEGFTTYAERRIVEVVQG 348
Query 235 AGVQRI------KEVADLISRQFAEDDGPMAHPIRPESYMAMDNFYTATVYDKGAEVVRI 288
A + + + + D + R +D+ + + D+ Y+ Y+KG + V
Sbjct 349 ADIATLNIGIGWRGLTDEMER--FKDNLECTKLWNKQEGVDPDDVYSQVPYEKGFQFVLR 406
Query 289 YHTLLGASGFRKGMDMYF 306
+G + F + + Y
Sbjct 407 IERQIGRTAFDEFLKKYI 424
> Hs13443031
Length=657
Score = 53.5 bits (127), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 88/352 (25%), Positives = 134/352 (38%), Gaps = 52/352 (14%)
Query 14 TQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVLLSNGNKTEEGLVHGSPDRHFAV 73
TQ +A R D P V KY +E + ++S + G P++ F
Sbjct 176 TQGQAVLNRAFFPCFDTPAVKYKYSALIEVPDG-FTAVMSARPGEKRG-----PNKFF-- 227
Query 74 FTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMHSVKVSMK 133
F P PSYL A+ AI D + + G V++EP + + ++ +
Sbjct 228 FQMCQPIPSYLIAL------AIGDLVSAEVGPRSR--VWAEPCLIDAAKEYNGVIEEFLA 279
Query 134 WEEEHFGR----EYDLDIFNVACVSDFNAGAMENKGLNIFNCTLLLADMQTSTDDDFERV 189
E+ FG YDL F G MEN L LL D R
Sbjct 280 TGEKLFGPYVWGRYDLLFMP----PSFPFGGMENPCLTFVTPCLLAGD----------RS 325
Query 190 LA-VVGHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGTVMSAGVQRIKEVAD--L 246
LA V+ HE H+W GN VT +W + L EG T++ ++ + T++ E A
Sbjct 326 LADVIIHEISHSWFGNLVTNANWGEFWLNEGFTMYAQRR-ISTILFGAAYTCLEAATGRA 384
Query 247 ISRQFAEDDG---PMAH-PIRPESYMAMDNFYTATVYDKGAEVVRIYHTLLG-ASGFRKG 301
+ RQ + G P+ ++ E + D+ Y T Y+KG V L+G F
Sbjct 385 LLRQHMDITGEENPLNKLRVKIEPGVDPDDTYNETPYEKGFCFVSYLAHLVGDQDQFDSF 444
Query 302 MDMYFERHDNTAATCDDF----RAAMADANRINLD-----QMDRWYSQAGTP 344
+ Y + DDF + + +D + DRW + G P
Sbjct 445 LKAYVHEFKFRSILADDFLDFYLEYFPELKKKRVDIIPGFEFDRWLNTPGWP 496
> Hs20537652
Length=650
Score = 52.8 bits (125), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 89/353 (25%), Positives = 136/353 (38%), Gaps = 53/353 (15%)
Query 14 TQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVLLSNGNKTEEGLVHGSPDRHFAV 73
TQ +A R D P V KY +E + ++S + G P++ F
Sbjct 168 TQGQAVLNRAFFPCFDTPAVKYKYSALIEVPDG-FTAVMSASTWEKRG-----PNKFF-- 219
Query 74 FTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMHSV-KVSM 132
F P PSYL A+ AI D + + G V++EP + + + V + +
Sbjct 220 FQMCQPIPSYLIAL------AIGDLVSAEVGPRSR--VWAEPCLIDAAKEEYNGVIEEFL 271
Query 133 KWEEEHFGR----EYDLDIFNVACVSDFNAGAMENKGLNIFNCTLLLADMQTSTDDDFER 188
E+ FG YDL + F G MEN L LL D R
Sbjct 272 ATGEKLFGPYVWGRYDL----LFMPPSFPFGGMENPCLTFVTPCLLAGD----------R 317
Query 189 VLA-VVGHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGTVMSAGVQRIKEVAD-- 245
LA V+ HE H+W GN VT +W + L EG T++ ++ + T++ E A
Sbjct 318 SLADVIIHEISHSWFGNLVTNANWGEFWLNEGFTMYAQRR-ISTILFGAAYTCLEAATGR 376
Query 246 LISRQFAEDDG---PMAH-PIRPESYMAMDNFYTATVYDKGAEVVRIYHTLLG-ASGFRK 300
+ RQ + G P+ ++ E + D+ Y T Y+KG V L+G F
Sbjct 377 ALLRQHMDITGEENPLNKLRVKIEPGVDPDDTYNETPYEKGFCFVSYLAHLVGDQDQFDS 436
Query 301 GMDMYFERHDNTAATCDDF----RAAMADANRINLD-----QMDRWYSQAGTP 344
+ Y + DDF + + +D + DRW + G P
Sbjct 437 FLKAYVHEFKFRSILADDFLDFYLEYFPELKKKRVDIIPGFEFDRWLNTPGWP 489
> Hs20070296
Length=494
Score = 51.6 bits (122), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 70/305 (22%), Positives = 116/305 (38%), Gaps = 42/305 (13%)
Query 59 EEGLVHGSPDRHFAVFTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQL 118
EEG+ H F HP P+YL A++AG+ ++ E L P
Sbjct 11 EEGVFH---------FHMEHPVPAYLVALVAGDLKPADIGPRSRVWAEPCLL----PTAT 57
Query 119 HKLEWAMHS-VKVSMKWEEEHFGREYDLDIFNVACVSDFNAGAMENKGLNIFNCTLLLAD 177
KL A+ + + + + YD+ V F AMEN L ++L +D
Sbjct 58 SKLSGAVEQWLSAAERLYGPYMWGRYDI----VFLPPSFPIVAMENPCLTFIISSILESD 113
Query 178 MQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTLKEGLTVFREQLFMGTVMSAGV 237
E ++ V HE H+W GN VT W ++ L EGL + ++ A
Sbjct 114 ---------EFLVIDVIHEVAHSWFGNAVTNATWEEMWLSEGLATYAQRRITTETYGAAF 164
Query 238 QRIKEV--ADLISRQ--FAEDDGPMAH-PIRPESYMAMDNFYTATVYDKGAEVVRIYHTL 292
++ D + RQ +D P++ ++ E + + Y+KG V L
Sbjct 165 TCLETAFRLDALHRQMKLLGEDSPVSKLQVKLEPGVNPSHLMNLFTYEKGYCFVYYLSQL 224
Query 293 LG-ASGFRKGMDMYFERHDNTAATC----DDFRAAMADANRINLD-----QMDRWYSQAG 342
G F + Y E++ T+ D F + + ++D + +RW + G
Sbjct 225 CGDPQRFDDFLRAYVEKYKFTSVVAQDLLDSFLSFFPELKEQSVDCRAGLEFERWLNATG 284
Query 343 TPRLE 347
P E
Sbjct 285 PPLAE 289
> YIL137c
Length=946
Score = 50.1 bits (118), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 54/307 (17%), Positives = 117/307 (38%), Gaps = 14/307 (4%)
Query 11 ALVTQCEAEGFRRITYFLDRPDVLAKYKVRLEANKAKYPVLLSNGNKTEEGLVHGSPDRH 70
+ T C+ I +D P + +++ + A A+Y + + + E L S +H
Sbjct 149 VVATHCQPFSASNIFPCIDEPSNKSTFQLNI-ATDAQYKAVSNTPVEMVEAL--DSSQKH 205
Query 71 FAVFTDPHPKPSYLFAILAGEFAAIRDKFTTKSGNEVALAVYSEPAQLHKLEWAMHSVKV 130
F + +F G+ ++ + + + +++Y+ P + + + +V+
Sbjct 206 LVKFAKTPLMTTSVFGFSIGDLEFLKTEIKLEGDRTIPVSIYA-PWDIANAAFTLDTVQK 264
Query 131 SMKWEEEHFGREYDLDIFNVACVSDFNAGAMENKGLNIFNCT-LLLADMQTSTDDDFERV 189
+ E +F Y L + + + AMEN G+ LL+ + + E+
Sbjct 265 YLPLLESYFKCPYPLPKLDFVLLPYLSDMAMENFGMITIQLNHLLIPPNALANETVREQA 324
Query 190 LAVVGHEYFHNWTGNRVTVRDWFQLTLKEGLT------VFREQLFMGTVMSAGVQRIKEV 243
++ HE H W GN ++ W L E + + + ++ +++V
Sbjct 325 QQLIVHELVHQWMGNYISFDSWESLWFNESFATWLACHILEQNGDLSHYWTSEPYLLQQV 384
Query 244 ADLISRQFAEDDGPMAHPIRPESY---MAMDNFYTATVYDKGAEVVRIYHTLLGASGFRK 300
+ R A+ +G I + + + Y KG ++R G S +K
Sbjct 385 EPTMCRDAADVNGRSIFQIAQRNTGIDSQTSDIFDPEAYTKGIIMLRSLQLATGESHLQK 444
Query 301 GMDMYFE 307
G++ FE
Sbjct 445 GLESVFE 451
Lambda K H
0.321 0.135 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8769821500
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40