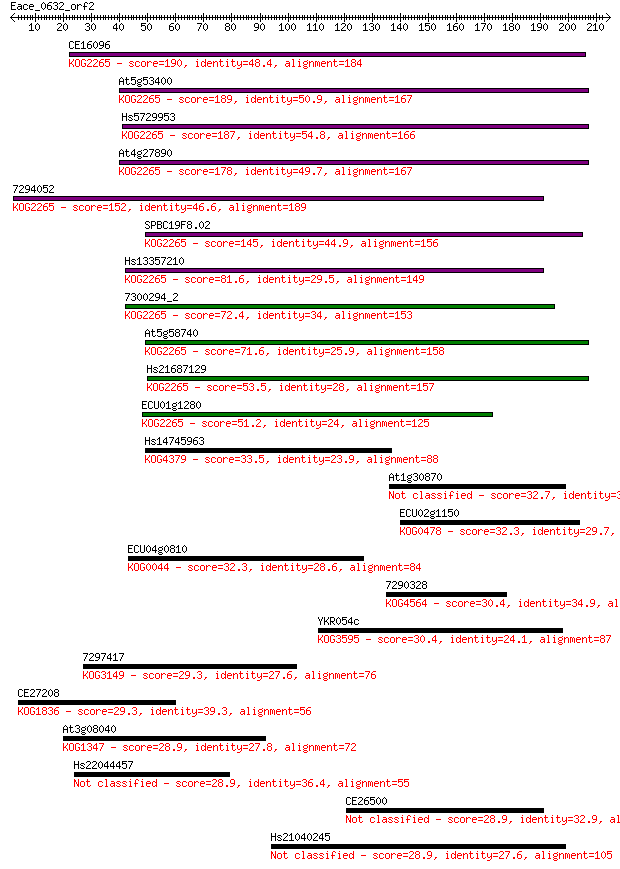
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0632_orf2
Length=214
Score E
Sequences producing significant alignments: (Bits) Value
CE16096 190 1e-48
At5g53400 189 2e-48
Hs5729953 187 2e-47
At4g27890 178 6e-45
7294052 152 4e-37
SPBC19F8.02 145 7e-35
Hs13357210 81.6 1e-15
7300294_2 72.4 6e-13
At5g58740 71.6 1e-12
Hs21687129 53.5 3e-07
ECU01g1280 51.2 1e-06
Hs14745963 33.5 0.34
At1g30870 32.7 0.58
ECU02g1150 32.3 0.73
ECU04g0810 32.3 0.77
7290328 30.4 2.7
YKR054c 30.4 3.1
7297417 29.3 5.8
CE27208 29.3 7.0
At3g08040 28.9 7.2
Hs22044457 28.9 7.4
CE26500 28.9 8.5
Hs21040245 28.9 9.1
> CE16096
Length=320
Score = 190 bits (483), Expect = 1e-48, Method: Compositional matrix adjust.
Identities = 89/187 (47%), Positives = 132/187 (70%), Gaps = 3/187 (1%)
Query 22 EKGEEGSTSDDDNESSRPE-GNGGQTDKYRWTQTLESVDVYVPMGEG--VRAAQCQVKIT 78
E ++ D+D++ +P GNG KY+WTQTL+ ++V +P+ G +++ VKI
Sbjct 133 ETSKDAEVEDEDSKLMKPNSGNGADLAKYQWTQTLQELEVKIPIAAGFAIKSRDVVVKIE 192
Query 79 ATSLTLGLKGQPPILNGEFSQRVQAEDSMWTLEDRKTLHLSLEKTDKMRWWSCVLKGDPE 138
TS+++GLK Q PI++G+ ++ E+ W +E+ K + L+LEK + M WW+ L DP
Sbjct 193 KTSVSVGLKNQAPIVDGKLPHAIKVENCNWVIENGKAIVLTLEKINDMEWWNRFLDSDPP 252
Query 139 IDTKTIVPENSKLSDLDPETRATVEKMMYDQQQKQKGLPTSDQQRQAEMLEKFKQAHPEL 198
I+TK + PENSKLSDLD ETRA VEKMMYDQ+QK+ GLPTSD++++ +ML++F + HPE+
Sbjct 253 INTKEVKPENSKLSDLDGETRAMVEKMMYDQRQKEMGLPTSDEKKKHDMLQQFMKQHPEM 312
Query 199 DFSNAKI 205
DFSNAKI
Sbjct 313 DFSNAKI 319
> At5g53400
Length=304
Score = 189 bits (481), Expect = 2e-48, Method: Compositional matrix adjust.
Identities = 85/167 (50%), Positives = 118/167 (70%), Gaps = 0/167 (0%)
Query 40 EGNGGQTDKYRWTQTLESVDVYVPMGEGVRAAQCQVKITATSLTLGLKGQPPILNGEFSQ 99
+GNG + Y W Q L+ V V +P+ G +A +I L +GLKGQ PI++GE +
Sbjct 138 KGNGTDLENYSWIQNLQEVTVNIPVPTGTKARTVVCEIKKNRLKVGLKGQDPIVDGELYR 197
Query 100 RVQAEDSMWTLEDRKTLHLSLEKTDKMRWWSCVLKGDPEIDTKTIVPENSKLSDLDPETR 159
V+ +D W +ED+K + + L K+D+M WW C +KG+PEIDT+ + PE SKL DLDPETR
Sbjct 198 SVKPDDCYWNIEDQKVISILLTKSDQMEWWKCCVKGEPEIDTQKVEPETSKLGDLDPETR 257
Query 160 ATVEKMMYDQQQKQKGLPTSDQQRQAEMLEKFKQAHPELDFSNAKIN 206
+TVEKMM+DQ+QKQ GLPTS++ ++ E+L+KF HPE+DFSNAK N
Sbjct 258 STVEKMMFDQRQKQMGLPTSEELQKQEILKKFMSEHPEMDFSNAKFN 304
> Hs5729953
Length=331
Score = 187 bits (474), Expect = 2e-47, Method: Compositional matrix adjust.
Identities = 91/168 (54%), Positives = 113/168 (67%), Gaps = 2/168 (1%)
Query 41 GNGGQTDKYRWTQTLESVDVYVPMGEGVR--AAQCQVKITATSLTLGLKGQPPILNGEFS 98
GNG YRWTQTL +D+ VP R V I L +GLKGQP I++GE
Sbjct 164 GNGADLPNYRWTQTLSELDLAVPFCVNFRLKGKDMVVDIQRRHLRVGLKGQPAIIDGELY 223
Query 99 QRVQAEDSMWTLEDRKTLHLSLEKTDKMRWWSCVLKGDPEIDTKTIVPENSKLSDLDPET 158
V+ E+S W +ED K + + LEK +KM WWS ++ DPEI+TK I PENSKLSDLD ET
Sbjct 224 NEVKVEESSWLIEDGKVVTVHLEKINKMEWWSRLVSSDPEINTKKINPENSKLSDLDSET 283
Query 159 RATVEKMMYDQQQKQKGLPTSDQQRQAEMLEKFKQAHPELDFSNAKIN 206
R+ VEKMMYDQ+QK GLPTSD+Q++ E+L+KF HPE+DFS AK N
Sbjct 284 RSMVEKMMYDQRQKSMGLPTSDEQKKQEILKKFMDQHPEMDFSKAKFN 331
> At4g27890
Length=293
Score = 178 bits (452), Expect = 6e-45, Method: Compositional matrix adjust.
Identities = 83/167 (49%), Positives = 114/167 (68%), Gaps = 0/167 (0%)
Query 40 EGNGGQTDKYRWTQTLESVDVYVPMGEGVRAAQCQVKITATSLTLGLKGQPPILNGEFSQ 99
+GNG +KY W Q L+ V + +PM EG ++ +I L +GLKGQ I++GEF
Sbjct 127 KGNGLDFEKYSWGQNLQEVTINIPMPEGTKSRSVTCEIKKNRLKVGLKGQDLIVDGEFFN 186
Query 100 RVQAEDSMWTLEDRKTLHLSLEKTDKMRWWSCVLKGDPEIDTKTIVPENSKLSDLDPETR 159
V+ +D W +ED+K + + L K D+M WW +KG+PEIDT+ + PE SKL DLDPETR
Sbjct 187 SVKPDDCFWNIEDQKMISVLLTKQDQMEWWKYCVKGEPEIDTQKVEPETSKLGDLDPETR 246
Query 160 ATVEKMMYDQQQKQKGLPTSDQQRQAEMLEKFKQAHPELDFSNAKIN 206
A+VEKMM+DQ+QKQ GLP SD+ + +ML+KF +P +DFSNAK N
Sbjct 247 ASVEKMMFDQRQKQMGLPRSDEIEKKDMLKKFMAQNPGMDFSNAKFN 293
> 7294052
Length=332
Score = 152 bits (385), Expect = 4e-37, Method: Compositional matrix adjust.
Identities = 88/215 (40%), Positives = 127/215 (59%), Gaps = 26/215 (12%)
Query 2 ESKAAETPSTAAEGATASSKEKGEEGSTSDDDNESSRPE--------GNGGQTDKYRWTQ 53
E+K + +A G ++S G D+ES + E GNG + Y WTQ
Sbjct 118 ETKKRQQLLDSAGGEPSASNRDGISKPIEKVDDESDKSELGKLMPNAGNGCTLENYTWTQ 177
Query 54 TLESVDVY----------------VP--MGEGVRAAQCQVKITATSLTLGLKGQPPILNG 95
TLE V+V +P + G+RA + I SL +G+KGQ PI++G
Sbjct 178 TLEEVEVSPGTSLTPYFSYLAQLKIPFNLTFGLRARDLVISIGKKSLKVGIKGQTPIIDG 237
Query 96 EFSQRVQAEDSMWTLEDRKTLHLSLEKTDKMRWWSCVLKGDPEIDTKTIVPENSKLSDLD 155
E V+ E+S+W L+D KT+ ++L+K +KM WWS ++ DPEI T+ I PE+SKLSDLD
Sbjct 238 ELCGEVKTEESVWVLQDSKTVMITLDKINKMNWWSRLVTTDPEISTRKINPESSKLSDLD 297
Query 156 PETRATVEKMMYDQQQKQKGLPTSDQQRQAEMLEK 190
ETR+ VEKMMYDQ+QK+ GLPTS+ +++ ++LEK
Sbjct 298 GETRSMVEKMMYDQRQKELGLPTSEDRKKQDILEK 332
> SPBC19F8.02
Length=166
Score = 145 bits (365), Expect = 7e-35, Method: Compositional matrix adjust.
Identities = 70/158 (44%), Positives = 107/158 (67%), Gaps = 7/158 (4%)
Query 49 YRWTQTLESVDVYVPMGEGVRAAQCQVKITATSLTLGLKG--QPPILNGEFSQRVQAEDS 106
Y W QT+ VD+ + + +G RA QV ++ L + + + +L+G +++ ++S
Sbjct 11 YEWDQTIADVDIVIHVPKGTRAKSLQVDMSNHDLKIQINVPERKVLLSGPLEKQINLDES 70
Query 107 MWTLEDRKTLHLSLEKTDKMRWWSCVLKGDPEIDTKTIVPENSKLSDLDPETRATVEKMM 166
WT+E+++ L + LEK++KM WWSCV+KG P ID +I PENSKLSDLD ETRATVEKMM
Sbjct 71 TWTVEEQERLVIHLEKSNKMEWWSCVIKGHPSIDIGSIEPENSKLSDLDEETRATVEKMM 130
Query 167 YDQQQKQKGLPTSDQQRQAEMLEKFKQAHPELDFSNAK 204
+Q QK+ +D+Q++ ++L+ F + HPELDFSN +
Sbjct 131 LEQSQKR-----TDEQKRKDVLQNFMKQHPELDFSNVR 163
> Hs13357210
Length=220
Score = 81.6 bits (200), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 44/153 (28%), Positives = 88/153 (57%), Gaps = 6/153 (3%)
Query 42 NGGQTDKYRWTQTLESVDVYVPMGEGV-RAAQCQVKITATSLTLGL---KGQPPILNGEF 97
NG + Y W+Q ++V VP+ + V + Q V ++++S+ + + G+ ++ G+
Sbjct 42 NGAVRENYTWSQDYTDLEVRVPVPKHVVKGKQVSVALSSSSIRVAMLEENGERVLMEGKL 101
Query 98 SQRVQAEDSMWTLEDRKTLHLSLEKTDKMRWWSCVLKGDPEIDTKTIVPENSKLSDLDPE 157
+ ++ E S+W+LE K + ++L K + WW+ +L+G+ ID I E S ++ +D E
Sbjct 102 THKINTESSLWSLEPGKCVLVNLSKVGEY-WWNAILEGEEPIDIDKINKERS-MATVDEE 159
Query 158 TRATVEKMMYDQQQKQKGLPTSDQQRQAEMLEK 190
+A ++++ +D QK +G P S + + EML+K
Sbjct 160 EQAVLDRLTFDYHQKLQGKPQSHELKVHEMLKK 192
> 7300294_2
Length=188
Score = 72.4 bits (176), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 52/158 (32%), Positives = 87/158 (55%), Gaps = 8/158 (5%)
Query 42 NGGQTDKYRWTQTLESVDVYVPMGEGVRAAQ-CQVKITATSLTLGLKGQPP--ILNGEFS 98
NG + + W+QTL+ V+V + + + A+ + I A + + K P IL G S
Sbjct 13 NGDVFETHCWSQTLKDVEVQALLPKDHQTAKKLHISIQAQHIKVSSKHSPETIILEGNLS 72
Query 99 QRVQAEDSMWTLEDRKTLHLSLEKTDKMRWWSCVLKGDPEIDTKTIVPENSKLSDLDPET 158
QR++ ++++WT+ D+ L +S +K ++ WW + +GDPEID+K I E + DL ET
Sbjct 73 QRIKHKEAVWTI-DQNRLIISYDKAKEL-WWDRLFEGDPEIDSKKIECERY-IDDLPEET 129
Query 159 RATVEKMMYDQ--QQKQKGLPTSDQQRQAEMLEKFKQA 194
+AT+EK+ Q KQ+ + QA L++ K A
Sbjct 130 QATIEKLRVQQLAADKQQNEIQTSSPDQAINLDRLKAA 167
> At5g58740
Length=158
Score = 71.6 bits (174), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 41/158 (25%), Positives = 74/158 (46%), Gaps = 36/158 (22%)
Query 49 YRWTQTLESVDVYVPMGEGVRAAQCQVKITATSLTLGLKGQPPILNGEFSQRVQAEDSMW 108
+ W QTLE V++Y+ + V KI + + +G+KG PP LN + S V+ + S W
Sbjct 21 FEWDQTLEEVNMYITLPPNVHPKSFHCKIQSKHIEVGIKGNPPYLNHDLSAPVKTDCSFW 80
Query 109 TLEDRKTLHLSLEKTDKMRWWSCVLKGDPEIDTKTIVPENSKLSDLDPETRATVEKMMYD 168
TLED +H++L+K +K + W+ + G ++D
Sbjct 81 TLED-DIMHITLQKREKGQTWASPILGQGQLDPY-------------------------- 113
Query 169 QQQKQKGLPTSDQQRQAEMLEKFKQAHPELDFSNAKIN 206
+D +++ ML++F++ +P DFS A+ +
Sbjct 114 ---------ATDLEQKRLMLQRFQEENPGFDFSQAQFS 142
> Hs21687129
Length=157
Score = 53.5 bits (127), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 44/159 (27%), Positives = 69/159 (43%), Gaps = 38/159 (23%)
Query 50 RWTQTLESVDVYVPMGEGVRAAQCQVKITATSLTLGLKGQPPILNGEFSQRVQAEDSMWT 109
+W QTLE V + V + G RA Q + + + L + G+ IL G+ A++ WT
Sbjct 20 QWYQTLEEVFIEVQVPPGTRAQDIQCGLQSRHVALSVGGRE-ILKGKLFDSTIADEGTWT 78
Query 110 LEDRKTLHLSLEKT--DKMRWWSCVLKGDPEIDTKTIVPENSKLSDLDPETRATVEKMMY 167
LEDRK + + L KT D W+ +L+ + D +
Sbjct 79 LEDRKMVRIVLTKTKRDAANCWTSLLESEYAADP-------------------------W 113
Query 168 DQQQKQKGLPTSDQQRQAEMLEKFKQAHPELDFSNAKIN 206
Q Q Q+ L LE+F++ +P DFS A+I+
Sbjct 114 VQDQMQRKL----------TLERFQKENPGFDFSGAEIS 142
> ECU01g1280
Length=131
Score = 51.2 bits (121), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 30/126 (23%), Positives = 64/126 (50%), Gaps = 4/126 (3%)
Query 48 KYRWTQTLESVDVYVPMGEGVRAAQCQVKITATSLTLGLKGQPPILNGEFSQRVQAEDSM 107
KY W Q L +++ P+ ++ +++I + + +G+ +++GE V S+
Sbjct 6 KYTWDQELNEINIQFPVTGDADSSAIKIRIVGKKICVKNQGEI-VIDGELLHEVDVS-SL 63
Query 108 WTLEDRKTLHLSLEKTDKMRWWSCVLKGDPEIDTKTIVP-ENSKLSDLDPETRATVEKMM 166
W + + + +++ K + WW +L G +D + + +++ +S LD E R VEKMM
Sbjct 64 WWVINGDVVDVNVTKK-RNEWWDSLLVGSESVDVQKLAENKHADMSMLDAEAREVVEKMM 122
Query 167 YDQQQK 172
++ K
Sbjct 123 HNTSGK 128
> Hs14745963
Length=583
Score = 33.5 bits (75), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 21/88 (23%), Positives = 40/88 (45%), Gaps = 1/88 (1%)
Query 49 YRWTQTLESVDVYVPMGEGVRAAQCQVKITATSLTLGLKGQPPILNGEFSQRVQAEDSMW 108
Y W QT + + V + + E Q++ + + LK L G+ + E S W
Sbjct 278 YYWQQTEDDLTVTIRLPEDSTKEDIQIQFLPDHINIVLKDHQ-FLEGKLYSSIDHESSTW 336
Query 109 TLEDRKTLHLSLEKTDKMRWWSCVLKGD 136
+++ +L +SL K ++ W ++ GD
Sbjct 337 IIKESNSLEISLIKKNEGLTWPELVIGD 364
> At1g30870
Length=349
Score = 32.7 bits (73), Expect = 0.58, Method: Compositional matrix adjust.
Identities = 22/74 (29%), Positives = 34/74 (45%), Gaps = 11/74 (14%)
Query 136 DPEIDTKTI------VPENSKLSDLDPETRATVEKMMYDQQQKQKGLPTSDQQ-----RQ 184
DP ID K S+ DLDP T A + Y QK G+ ++DQ+ R
Sbjct 237 DPSIDAKYADYLQRRCRWASETVDLDPVTPAVFDNQYYINLQKHMGVLSTDQELVKDPRT 296
Query 185 AEMLEKFKQAHPEL 198
A +++ F + P++
Sbjct 297 APLVKTFAEQSPQI 310
> ECU02g1150
Length=708
Score = 32.3 bits (72), Expect = 0.73, Method: Composition-based stats.
Identities = 19/64 (29%), Positives = 29/64 (45%), Gaps = 6/64 (9%)
Query 140 DTKTIVPENSKLSDLDPETRATVEKMMYDQQQKQKGLPTSDQQRQAEMLEKFKQAHPELD 199
+ + IVP L PE+ + + D +Q G + RQ E L + +AH +
Sbjct 547 EARRIVPR------LTPESMKMLTQSYVDLRQMDNGKTITATTRQLESLIRLSEAHARMR 600
Query 200 FSNA 203
FSNA
Sbjct 601 FSNA 604
> ECU04g0810
Length=184
Score = 32.3 bits (72), Expect = 0.77, Method: Compositional matrix adjust.
Identities = 24/86 (27%), Positives = 37/86 (43%), Gaps = 4/86 (4%)
Query 43 GGQTDKYRWTQTL--ESVDVYVPMGEGVRAAQCQVKITATSLTLGLKGQPPILNGEFSQR 100
G +K RW + D V E V QC +T+ +L + + + + E
Sbjct 95 GSDHEKLRWMFRFYDKDNDGVVSKEEMVEVVQCLADMTSHTLDISIDAEGIV--SEIFAS 152
Query 101 VQAEDSMWTLEDRKTLHLSLEKTDKM 126
V+++ T ED KTL + EK KM
Sbjct 153 VESDRGFLTFEDFKTLSIRSEKLFKM 178
> 7290328
Length=633
Score = 30.4 bits (67), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 15/43 (34%), Positives = 25/43 (58%), Gaps = 2/43 (4%)
Query 135 GDPEIDTKTIVPENSKLSDLDPETRATVEKMMYDQQQKQKGLP 177
GDP K I P N +L+ P + + V M+++ QQ+Q+ +P
Sbjct 514 GDPSATGKRISPPNKRLNGRKPSSASIV--MIHEPQQRQRLMP 554
> YKR054c
Length=4092
Score = 30.4 bits (67), Expect = 3.1, Method: Composition-based stats.
Identities = 21/87 (24%), Positives = 52/87 (59%), Gaps = 13/87 (14%)
Query 111 EDRKTLHLSLEKTDKMRWWSCVLKGDPEIDTKTIVPENSKLSDLDPETRATVEKMMYDQQ 170
E+++ +++ LEK ++ VLK + E++ KT+ ++++L++ + E R+T++KM+ +Q
Sbjct 3012 ENQRFVNVGLEKLNE-----SVLKVN-ELN-KTLSKKSTELTEKEKEARSTLDKMLMEQN 3064
Query 171 QKQKGLPTSDQQRQAEMLEKFKQAHPE 197
+ ++ +Q E ++K + E
Sbjct 3065 ESER------KQEATEEIKKILKVQEE 3085
> 7297417
Length=969
Score = 29.3 bits (64), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 21/81 (25%), Positives = 37/81 (45%), Gaps = 5/81 (6%)
Query 27 GSTSDDDNESSRPEGNGGQTDKYRWTQTLESVDVYVPMGEGVRAAQCQVKITAT-SLTLG 85
G+TS E SR G GG Y+W ++ D+ P+ + ++ + + + + +
Sbjct 254 GNTSKYIGEDSRENGTGGNALTYKWLVYVQGKDLPEPLEKYIKKVRFHLHPSYRPNDIVD 313
Query 86 LKGQPPILN----GEFSQRVQ 102
+ P LN GEF R+Q
Sbjct 314 VHRSPFQLNRHGWGEFPMRIQ 334
> CE27208
Length=2760
Score = 29.3 bits (64), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 22/56 (39%), Positives = 28/56 (50%), Gaps = 4/56 (7%)
Query 4 KAAETPSTAAEGATASSKEKGEEGSTSDDDNESSRPEGNGGQTDKYRWTQTLESVD 59
A E +AAE + A SK G EGS S D NE S Q +K + +L +VD
Sbjct 2048 NATEAVDSAAEASEAVSKMLGSEGSESGDANEESLRS----QLEKLKNESSLSNVD 2099
> At3g08040
Length=526
Score = 28.9 bits (63), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 20/77 (25%), Positives = 34/77 (44%), Gaps = 5/77 (6%)
Query 20 SKEKGEEGSTSDDDNESSRP-----EGNGGQTDKYRWTQTLESVDVYVPMGEGVRAAQCQ 74
S EKG TS+D N+ +P + N G + +T+ + + +G + Q
Sbjct 128 SLEKGISSPTSNDTNQPQQPPAPDTKSNSGNKSNKKEKRTIRTASTAMILGLILGLVQAI 187
Query 75 VKITATSLTLGLKGQPP 91
I ++ L LG+ G P
Sbjct 188 FLIFSSKLLLGVMGVKP 204
> Hs22044457
Length=170
Score = 28.9 bits (63), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 20/58 (34%), Positives = 29/58 (50%), Gaps = 5/58 (8%)
Query 24 GEEGSTSDDDNESSRPEGNGGQTDKYRWTQTLESVDVYV--PMGEGV-RAAQCQVKIT 78
GE D D+ +RP G G++ WTQ L ++ P+ EG+ RA C + IT
Sbjct 106 GESAGAEDSDDPETRPNGEKGESK--LWTQKLFTLMYKTSKPIDEGLSRAEPCYLLIT 161
> CE26500
Length=668
Score = 28.9 bits (63), Expect = 8.5, Method: Composition-based stats.
Identities = 23/71 (32%), Positives = 36/71 (50%), Gaps = 7/71 (9%)
Query 121 EKTDKM-RWWSCVLKGDPEIDTKTIVPENSKLSDLDPETRATVEKMMYDQQQKQKGLPTS 179
EK KM +W LK D +D+K ++ DL E R + K++ +Q + T+
Sbjct 171 EKCLKMLEFW---LKTDQNLDSKA---KSLIFEDLSLENRPKIGKLIALEQALTEAQDTN 224
Query 180 DQQRQAEMLEK 190
D R+A +LEK
Sbjct 225 DMYREAGILEK 235
> Hs21040245
Length=445
Score = 28.9 bits (63), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 29/110 (26%), Positives = 49/110 (44%), Gaps = 11/110 (10%)
Query 94 NGEFSQRVQAEDSMWTLEDRKTLHLSL--EKTDKMRWWSCVLKGDPEIDTKTIVPENSKL 151
N F + + + +R+ + ++L E D+ + W + EI K + +
Sbjct 74 NKSFYEVINVSPGYQLVRNREQISVTLGDEMFDRKKRW------ESEIPDKGRFSRTNII 127
Query 152 SDLD---PETRATVEKMMYDQQQKQKGLPTSDQQRQAEMLEKFKQAHPEL 198
SDL+ E A +E+M D Q QK L + R AEM + F+ + EL
Sbjct 128 SDLEEQISELTAIIEQMNRDHQSAQKLLSSEMDLRCAEMKQNFENKNREL 177
Lambda K H
0.306 0.125 0.363
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3933248660
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40