bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0711_orf1
Length=156
Score E
Sequences producing significant alignments: (Bits) Value
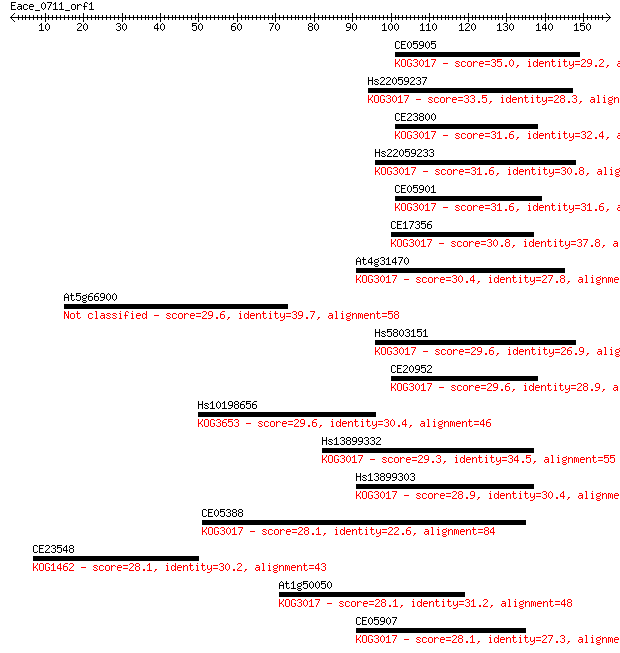
CE05905 35.0 0.052
Hs22059237 33.5 0.18
CE23800 31.6 0.58
Hs22059233 31.6 0.70
CE05901 31.6 0.74
CE17356 30.8 1.0
At4g31470 30.4 1.3
At5g66900 29.6 2.2
Hs5803151 29.6 2.4
CE20952 29.6 2.4
Hs10198656 29.6 2.6
Hs13899332 29.3 3.1
Hs13899303 28.9 4.1
CE05388 28.1 6.8
CE23548 28.1 7.2
At1g50050 28.1 7.2
CE05907 28.1 7.7
> CE05905
Length=207
Score = 35.0 bits (79), Expect = 0.052, Method: Compositional matrix adjust.
Identities = 14/48 (29%), Positives = 23/48 (47%), Gaps = 0/48 (0%)
Query 101 LLWSESTKIACAVGVCTDDTPLPSGKEAILVCQFSPAAQENAAPFSKE 148
+ W+ ++ I C V C D + + + +VCQ+SP P KE
Sbjct 139 MAWANTSSIGCGVKNCGRDASMRNMNKIAVVCQYSPPGNTMGRPIYKE 186
> Hs22059237
Length=344
Score = 33.5 bits (75), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 15/53 (28%), Positives = 24/53 (45%), Gaps = 2/53 (3%)
Query 94 DGANLANLLWSESTKIACAVGVCTDDTPLPSGKEAILVCQFSPAAQENAAPFS 146
D +N L+W S K+ CAV C+ + AI +C ++P P+
Sbjct 154 DCSNYIQLVWDHSYKVGCAVTPCSKIGHIIHA--AIFICNYAPGGTLTRRPYE 204
> CE23800
Length=208
Score = 31.6 bits (70), Expect = 0.58, Method: Compositional matrix adjust.
Identities = 12/37 (32%), Positives = 21/37 (56%), Gaps = 0/37 (0%)
Query 101 LLWSESTKIACAVGVCTDDTPLPSGKEAILVCQFSPA 137
+ W+E+ KI C V C D+ + + + +VCQ+ A
Sbjct 140 MAWAETNKIGCGVKNCGKDSSMNNMYKVAVVCQYDQA 176
> Hs22059233
Length=233
Score = 31.6 bits (70), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 16/53 (30%), Positives = 25/53 (47%), Gaps = 4/53 (7%)
Query 96 ANLANLLWSESTKIACAVGVCTDDTPLPSGKEAILVCQFSPAAQ-ENAAPFSK 147
+ L+W+ S + CAV +C + L AI VC + PA N P+ +
Sbjct 136 GHYTQLVWANSFYVGCAVAMCPN---LGGASTAIFVCNYGPAGNFANMPPYVR 185
> CE05901
Length=212
Score = 31.6 bits (70), Expect = 0.74, Method: Compositional matrix adjust.
Identities = 12/38 (31%), Positives = 22/38 (57%), Gaps = 3/38 (7%)
Query 101 LLWSESTKIACAVGVCTDDTPLPSGKEAILVCQFSPAA 138
+ W+ + KI C + C+ D+ G + ++VC +SPA
Sbjct 147 MAWATTNKIGCGISKCSSDS---FGTQYVVVCLYSPAG 181
> CE17356
Length=241
Score = 30.8 bits (68), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 20/39 (51%), Gaps = 2/39 (5%)
Query 100 NLLWSESTKIACAVGVC--TDDTPLPSGKEAILVCQFSP 136
+LW E+ K+ CAV C D L K + VC++ P
Sbjct 155 QILWKETRKLGCAVQECPARQDGSLDGQKYNVAVCKYYP 193
> At4g31470
Length=185
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 15/55 (27%), Positives = 25/55 (45%), Gaps = 9/55 (16%)
Query 91 SNPDGANLANLLWSESTKIACAVGVC-TDDTPLPSGKEAILVCQFSPAAQENAAP 144
+N D + L+W +S++I CA+ C T DT ++C + P P
Sbjct 137 ANGDCLHYTQLVWKKSSRIGCAISFCKTGDT--------FIICNYDPPGNIVGQP 183
> At5g66900
Length=809
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 23/61 (37%), Positives = 30/61 (49%), Gaps = 8/61 (13%)
Query 15 DDMVKTLKTALGDGTGLST--SDTCDNMALGASLK-LTFNVKFTQETSKTPNYRDMVQTA 71
DD V TL + G G +T S CD+ + K + FNV S TPN+R +VQ
Sbjct 184 DDSVVTLVVSAPPGCGKTTLVSRLCDDPDIKGKFKHIFFNV-----VSNTPNFRVIVQNL 238
Query 72 L 72
L
Sbjct 239 L 239
> Hs5803151
Length=266
Score = 29.6 bits (65), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 14/57 (24%), Positives = 26/57 (45%), Gaps = 9/57 (15%)
Query 96 ANLANLLWSESTKIACAVGVCTDDTPLPSGKEAI-----LVCQFSPAAQENAAPFSK 147
+ ++W++S K+ CAV C P SG +A+ +C + P P+ +
Sbjct 136 GHYTQVVWADSYKVGCAVQFC----PKVSGFDALSNGAHFICNYGPGGNYPTWPYKR 188
> CE20952
Length=208
Score = 29.6 bits (65), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 11/38 (28%), Positives = 20/38 (52%), Gaps = 0/38 (0%)
Query 100 NLLWSESTKIACAVGVCTDDTPLPSGKEAILVCQFSPA 137
+ W+E++KI C + C D + + +VCQ+ A
Sbjct 139 QMAWAETSKIGCGIKNCGKDANKKNMYKVAVVCQYDSA 176
> Hs10198656
Length=573
Score = 29.6 bits (65), Expect = 2.6, Method: Composition-based stats.
Identities = 14/46 (30%), Positives = 23/46 (50%), Gaps = 2/46 (4%)
Query 50 FNVKFTQETSKTPNYRDMVQTALNKGLGQLPTYPSTWNAFWSNPDG 95
F + + E TP ++ A+ + + P PSTW F ++PDG
Sbjct 436 FQLAYEAELGNTPTSDELWALAVQER--RRPYIPSTWRCFATDPDG 479
> Hs13899332
Length=497
Score = 29.3 bits (64), Expect = 3.1, Method: Composition-based stats.
Identities = 19/61 (31%), Positives = 26/61 (42%), Gaps = 6/61 (9%)
Query 82 YPSTWNAFW----SNPDGANLANLLWSESTKIACAVGVCTDDTPLPSGKE--AILVCQFS 135
YPS N + S P + ++W+ + KI CAV C T E VC +S
Sbjct 142 YPSECNPWCPERCSGPMCTHYTQIVWATTNKIGCAVNTCRKMTVWGEVWENAVYFVCNYS 201
Query 136 P 136
P
Sbjct 202 P 202
> Hs13899303
Length=500
Score = 28.9 bits (63), Expect = 4.1, Method: Composition-based stats.
Identities = 14/48 (29%), Positives = 23/48 (47%), Gaps = 2/48 (4%)
Query 91 SNPDGANLANLLWSESTKIACAVGVCTDDTPLPS--GKEAILVCQFSP 136
S P + ++W+ S +I CA+ +C + K LVC +SP
Sbjct 161 SGPVCTHYTQVVWATSNRIGCAINLCHNMNIWGQIWPKAVYLVCNYSP 208
> CE05388
Length=209
Score = 28.1 bits (61), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 19/90 (21%), Positives = 39/90 (43%), Gaps = 9/90 (10%)
Query 51 NVKFTQETSKTPNYRDMVQTALNKGLGQLPTYPSTWN------AFWSNPDGANLANLLWS 104
N+ + +S + VQ+A++ + + + WN A W+ G + + WS
Sbjct 88 NLFWAYSSSPITDLDKYVQSAVDTWVSEFQMFG--WNSNKFTTALWNTGIG-HATQVAWS 144
Query 105 ESTKIACAVGVCTDDTPLPSGKEAILVCQF 134
+ ++ C C D+ +A +VCQ+
Sbjct 145 ATGQVGCGAKNCGADSVRVGSYKATIVCQY 174
> CE23548
Length=404
Score = 28.1 bits (61), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 13/43 (30%), Positives = 22/43 (51%), Gaps = 0/43 (0%)
Query 7 RINEVTKADDMVKTLKTALGDGTGLSTSDTCDNMALGASLKLT 49
RI E TK + + +G+G +S S CD + +G + +T
Sbjct 330 RIGEKTKLKESIIAKGVVIGNGASISNSIICDGVEIGENADVT 372
> At1g50050
Length=226
Score = 28.1 bits (61), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 23/48 (47%), Gaps = 0/48 (0%)
Query 71 ALNKGLGQLPTYPSTWNAFWSNPDGANLANLLWSESTKIACAVGVCTD 118
A+N + + P Y T NA + ++WS S KI CA +C +
Sbjct 94 AVNLWVNEKPYYNYTANACIGAQQCKHYTQVVWSNSVKIGCARVLCNN 141
> CE05907
Length=211
Score = 28.1 bits (61), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 12/44 (27%), Positives = 21/44 (47%), Gaps = 0/44 (0%)
Query 91 SNPDGANLANLLWSESTKIACAVGVCTDDTPLPSGKEAILVCQF 134
+N + + W+ + I C V C D L + A++VCQ+
Sbjct 133 ANTGIGHATQMAWANTGLIGCGVKNCGPDPELNNYNRAVVVCQY 176
Lambda K H
0.313 0.127 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2070320142
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40