bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0741_orf1
Length=136
Score E
Sequences producing significant alignments: (Bits) Value
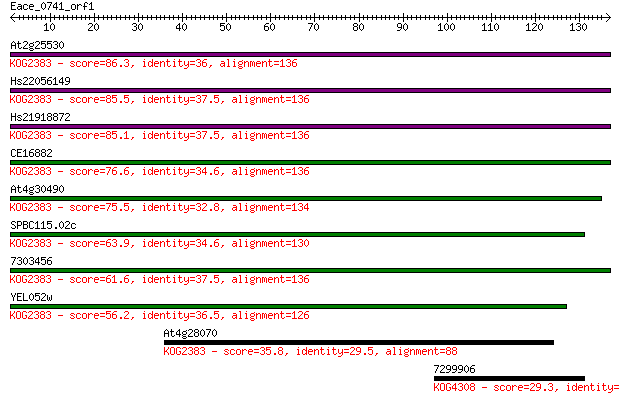
At2g25530 86.3 2e-17
Hs22056149 85.5 3e-17
Hs21918872 85.1 4e-17
CE16882 76.6 1e-14
At4g30490 75.5 3e-14
SPBC115.02c 63.9 8e-11
7303456 61.6 4e-10
YEL052w 56.2 2e-08
At4g28070 35.8 0.027
7299906 29.3 2.4
> At2g25530
Length=655
Score = 86.3 bits (212), Expect = 2e-17, Method: Composition-based stats.
Identities = 49/142 (34%), Positives = 77/142 (54%), Gaps = 6/142 (4%)
Query 1 GVVVVATSNRPPDDLYLGGLNRERFISFIPLLKTFCNIHHIETQKDYRLAKGAKSSGSLF 60
G V+VATSNR P +L G+ +E F FI L+ C I I ++ DYR S+ ++
Sbjct 386 GTVLVATSNRAPRELNQDGMQKEIFDKFISKLEKHCEIISIGSEVDYRRVAAKNSAENVH 445
Query 61 FV-PSRAK-----EIIFKQIEGAAGGSLESGSIPLAANRTLRVPGMRKGYAVFTFDDLCG 114
++ P E +++Q+ GG + S ++P+ RT+ VP G A FTF+ LCG
Sbjct 446 YLWPLNDAVLEEFEKMWRQVTDQYGGEITSATLPVMFGRTVEVPESCSGVARFTFEYLCG 505
Query 115 DNVGAVDYLYISSVLHSLSIRN 136
VGA DY+ ++ H++ I +
Sbjct 506 RPVGAADYIAVAKNYHTIFISD 527
> Hs22056149
Length=338
Score = 85.5 bits (210), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 51/141 (36%), Positives = 76/141 (53%), Gaps = 7/141 (4%)
Query 1 GVVVVATSNRPPDDLYLGGLNRERFISFIPLLKTFCNIHHIETQKDYRLAKGAKSSGSLF 60
GVVVVATSNRPP+DLY GL R F+ FI +LK +CN +++ DYR + ++G L+
Sbjct 94 GVVVVATSNRPPEDLYKNGLQRANFVPFIAVLKEYCNTVQLDSGIDYR-KRELPAAGKLY 152
Query 61 FVPSRAK-----EIIFKQIEGAAGGSLESGSIPLAANRTLRVPGMRKGYAVFTFDDLCGD 115
++ S A + +F ++ L I R LR+ A TF++LC
Sbjct 153 YLTSEADVEAVMDKLFDEL-AQKQNDLTRPRILKVQGRELRLNKACGTVADCTFEELCER 211
Query 116 NVGAVDYLYISSVLHSLSIRN 136
+GA DYL +S ++ +RN
Sbjct 212 PLGASDYLELSKNFDTIFLRN 232
> Hs21918872
Length=481
Score = 85.1 bits (209), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 51/141 (36%), Positives = 76/141 (53%), Gaps = 7/141 (4%)
Query 1 GVVVVATSNRPPDDLYLGGLNRERFISFIPLLKTFCNIHHIETQKDYRLAKGAKSSGSLF 60
GVVVVATSNRPP+DLY GL R F+ FI +LK +CN +++ DYR + ++G L+
Sbjct 237 GVVVVATSNRPPEDLYKNGLQRANFVPFIAVLKEYCNTVQLDSGIDYR-KRELPAAGKLY 295
Query 61 FVPSRAK-----EIIFKQIEGAAGGSLESGSIPLAANRTLRVPGMRKGYAVFTFDDLCGD 115
++ S A + +F ++ L I R LR+ A TF++LC
Sbjct 296 YLTSEADVEAVMDKLFDEL-AQKQNDLTRPRILKVQGRELRLNKACGTVADCTFEELCER 354
Query 116 NVGAVDYLYISSVLHSLSIRN 136
+GA DYL +S ++ +RN
Sbjct 355 PLGASDYLELSKNFDTIFLRN 375
> CE16882
Length=445
Score = 76.6 bits (187), Expect = 1e-14, Method: Composition-based stats.
Identities = 47/140 (33%), Positives = 73/140 (52%), Gaps = 7/140 (5%)
Query 1 GVVVVATSNRPPDDLYLGGLNRERFISFIPLLKTFCNIHHIETQKDYRLAKGAKSSGSLF 60
G+V+VATSNR P +LY GL R +F+ FI +L+ C +++ DYR + A G+
Sbjct 193 GLVMVATSNRAPSELYKNGLQRHQFMPFITILEDKCASLALDSGMDYR--RSAAGDGNHV 250
Query 61 FVPSRAK----EIIFKQIEGAAGGSLESGSIPLAANRTLRVPGMRKGYAVFTFDDLCGDN 116
+ S +I+FKQ ++ S ++ + R + V G A F +LC
Sbjct 251 YFSSEDSNTQCDIVFKQSAANENDTVRSKTLEILGRRVI-VEKCCGGVADVDFKELCMTA 309
Query 117 VGAVDYLYISSVLHSLSIRN 136
GAVDYL + V H++ +RN
Sbjct 310 KGAVDYLVYARVFHTVIVRN 329
> At4g30490
Length=447
Score = 75.5 bits (184), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 44/136 (32%), Positives = 70/136 (51%), Gaps = 4/136 (2%)
Query 1 GVVVVATSNRPPDDLYLGGLNRERFISFIPLLKTFCNIHHIETQKDYRLAKGAKSSGSLF 60
GV++VATSNR PD LY GGL R+ F+ FI LK +H I + DYR K + +
Sbjct 227 GVILVATSNRNPDKLYEGGLQRDLFLPFISSLKERSVVHEIGSAVDYR--KLTSAEQGFY 284
Query 61 FVPSRAKEIIFKQIEGAAGGSL--ESGSIPLAANRTLRVPGMRKGYAVFTFDDLCGDNVG 118
F+ ++ ++ G ++ + + R L++P G A F F++LC +G
Sbjct 285 FIGKDLSTLLKQKFRQLIGDNVVARPQVVEVVMGRKLQIPLGANGCAYFPFEELCDRPLG 344
Query 119 AVDYLYISSVLHSLSI 134
A DY + H+L++
Sbjct 345 AADYFGLFKKFHTLAL 360
> SPBC115.02c
Length=454
Score = 63.9 bits (154), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 45/143 (31%), Positives = 64/143 (44%), Gaps = 24/143 (16%)
Query 1 GVVVVATSNRPPDDLYLGGLNRERFISFIPLLKTFCNIHHIETQKDYRLAKGAKSSGSLF 60
GVV+ TSNR P DLY G+ RE FI I LL+ + +++ DYR K L+
Sbjct 220 GVVIFITSNRAPSDLYKNGIQRESFIPCIKLLEHRLQVICLDSPNDYRRLKSKTEDTYLY 279
Query 61 FVPSRAKEIIFKQIEGAAGGSLESGSIPLAAN-------------RTLRVPGMRKGYAVF 107
P+ + E+ +LE+ + A R + VP A F
Sbjct 280 --PANSPEV---------KKALENWFLCYADEKDPAHQDEVEVFGRKIIVPKASGNVAWF 328
Query 108 TFDDLCGDNVGAVDYLYISSVLH 130
TF+ LCG+ A DYL ++S H
Sbjct 329 TFEQLCGEPKSAADYLSLASRYH 351
> 7303456
Length=458
Score = 61.6 bits (148), Expect = 4e-10, Method: Composition-based stats.
Identities = 51/150 (34%), Positives = 71/150 (47%), Gaps = 27/150 (18%)
Query 1 GVVVVATSNRPPDDLYLGGLNRERFISFIPLLKTFCNIHHIETQKDYRLAKGAKSSGSLF 60
G+VVVATSNR P+DLY GL R F+ FI LL+ C + +++ DYR + A+S + +
Sbjct 219 GIVVVATSNRHPEDLYKNGLQRTNFLPFIALLQRRCKVSQLDS-IDYR--RIAQSGDTNY 275
Query 61 FVPSRAKEIIFKQIEGAAGGSLESGSIPLAAN-------RTLRVPGM-----RKGYAVF- 107
FV + A GS+ L A RTL G R V
Sbjct 276 FVKGQTD----------ADGSMNRMFKILCAEENDIIRPRTLTHFGRDLTFNRTCGQVLD 325
Query 108 -TFDDLCGDNVGAVDYLYISSVLHSLSIRN 136
TF +LC + D+L IS H++ IR+
Sbjct 326 STFSELCDRPLAGSDFLQISQFFHTVLIRD 355
> YEL052w
Length=509
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 46/153 (30%), Positives = 69/153 (45%), Gaps = 29/153 (18%)
Query 1 GVVVVATSNRPPDDLYLGGLNRERFISFIPLLKTFCNIHHIETQKDYRLAKGAKSSGSLF 60
GVV+ ATSNR PD+LY+ G+ R+ FI I L+K + + + DYR K + S++
Sbjct 231 GVVLFATSNRHPDELYINGVQRQSFIPCIELIKHRTKVIFLNSPTDYR--KIPRPVSSVY 288
Query 61 FVPSRA------------KEIIFKQI------EGAAGGSLESGSI-------PLA-ANRT 94
+ PS +E K+ S +S ++ PL R
Sbjct 289 YFPSDTSIKYASKECKTRRETHIKEWYNYFAQASHTDDSTDSHTVHKTFYDYPLTIWGRE 348
Query 95 LRVPGMRKG-YAVFTFDDLCGDNVGAVDYLYIS 126
+VP A FTF LCG+ + A DYL ++
Sbjct 349 FKVPKCTPPRVAQFTFKQLCGEPLAAGDYLTLA 381
> At4g28070
Length=345
Score = 35.8 bits (81), Expect = 0.027, Method: Compositional matrix adjust.
Identities = 26/90 (28%), Positives = 39/90 (43%), Gaps = 5/90 (5%)
Query 36 CNIHHIETQKDYRLAKGAKSSGSLFFVPSRAKEIIFKQIEGAAGGSLESGS--IPLAANR 93
C + I + DYR A+ F+ + + KQ G +G + + R
Sbjct 189 CVVREIGSSVDYRKLTSAEEG---FYFIGKDISGLLKQKFQLLVGDQPAGPQVVEVVMGR 245
Query 94 TLRVPGMRKGYAVFTFDDLCGDNVGAVDYL 123
L+VP G A F F++LC +GA DYL
Sbjct 246 KLQVPLAADGCAYFLFEELCDRPLGAADYL 275
> 7299906
Length=811
Score = 29.3 bits (64), Expect = 2.4, Method: Composition-based stats.
Identities = 14/35 (40%), Positives = 20/35 (57%), Gaps = 1/35 (2%)
Query 97 VPGMR-KGYAVFTFDDLCGDNVGAVDYLYISSVLH 130
VP +R K A TF +LCGD + D+ ++ LH
Sbjct 50 VPEIRSKSNATTTFLELCGDKLAVSDWQLLTEALH 84
Lambda K H
0.322 0.140 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1425342594
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40