bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0760_orf2
Length=80
Score E
Sequences producing significant alignments: (Bits) Value
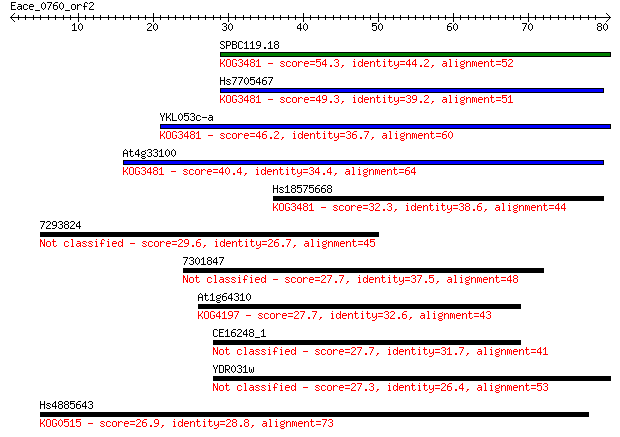
SPBC119.18 54.3 6e-08
Hs7705467 49.3 2e-06
YKL053c-a 46.2 2e-05
At4g33100 40.4 9e-04
Hs18575668 32.3 0.21
7293824 29.6 1.3
7301847 27.7 5.3
At1g64310 27.7 5.7
CE16248_1 27.7 6.0
YDR031w 27.3 7.6
Hs4885643 26.9 9.1
> SPBC119.18
Length=69
Score = 54.3 bits (129), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 23/53 (43%), Positives = 37/53 (69%), Gaps = 1/53 (1%)
Query 29 DTCSGLKEQYDRCFNHWYRRHFLRGDM-SRNCFHLFADYRLCISEELKRKGLE 80
+ C+ K++YD CFN WY FL+GD+ +R+C LFA+Y+ C+ + LK K ++
Sbjct 7 EECTPAKKKYDACFNDWYANKFLKGDLHNRDCDELFAEYKSCLLKALKTKKID 59
> Hs7705467
Length=76
Score = 49.3 bits (116), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 20/52 (38%), Positives = 33/52 (63%), Gaps = 1/52 (1%)
Query 29 DTCSGLKEQYDRCFNHWYRRHFLRGDMSRN-CFHLFADYRLCISEELKRKGL 79
+ C+ +K +YD+CFN W+ FL+GD S + C LF Y+ C+ + +K K +
Sbjct 6 EACTDMKREYDQCFNRWFAEKFLKGDSSGDPCTDLFKRYQQCVQKAIKEKEI 57
> YKL053c-a
Length=86
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 22/61 (36%), Positives = 35/61 (57%), Gaps = 4/61 (6%)
Query 21 AAACAPQMDTCSGLKEQYDRCFNHWYRRHFLRGDMSRN-CFHLFADYRLCISEELKRKGL 79
+A+ AP+ C+ LK +YD CFN WY FL+G N C + Y C++ L ++G+
Sbjct 6 SASFAPE---CTDLKTKYDSCFNEWYSEKFLKGKSVENECSKQWYAYTTCVNAALVKQGI 62
Query 80 E 80
+
Sbjct 63 K 63
> At4g33100
Length=92
Score = 40.4 bits (93), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 22/65 (33%), Positives = 33/65 (50%), Gaps = 5/65 (7%)
Query 16 DFWTAAAACAPQMDTCSGLKEQYDRCFNHWYRRHFLRGDMSR-NCFHLFADYRLCISEEL 74
D +A ++ +P C+ L+ Y CFN WY F++G + C + YR C+SE L
Sbjct 8 DSTSARSSTSP----CADLRNAYHNCFNKWYSEKFVKGQWDKEECVAEWKKYRDCLSENL 63
Query 75 KRKGL 79
K L
Sbjct 64 DGKLL 68
> Hs18575668
Length=91
Score = 32.3 bits (72), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 27/49 (55%), Gaps = 5/49 (10%)
Query 36 EQYDRCFNHWYRRHFLRGDMSRN-CFHLFADYRLCI----SEELKRKGL 79
+ YD+CF+ W+ L+GD S C +LF Y C+ +E + +GL
Sbjct 14 QSYDQCFSCWFVEKLLKGDSSGEPCSNLFKHYLQCVQKATTERISVEGL 62
> 7293824
Length=190
Score = 29.6 bits (65), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 12/46 (26%), Positives = 19/46 (41%), Gaps = 1/46 (2%)
Query 5 SGIAPSAYVDADFWTAAAACAPQMDT-CSGLKEQYDRCFNHWYRRH 49
G+ + + W QM T C L E+Y+ + WY +H
Sbjct 127 KGVKVDLGIPYELWDKPPVEVTQMKTQCENLLEEYEETISEWYFKH 172
> 7301847
Length=631
Score = 27.7 bits (60), Expect = 5.3, Method: Composition-based stats.
Identities = 18/51 (35%), Positives = 26/51 (50%), Gaps = 8/51 (15%)
Query 24 CAPQMDTCSGLK-EQYDRCFNHWYRRHFLRGDMS--RNCFHLFADYRLCIS 71
CA CS ++ E Y+ C RH LR S R C H++++ R CI+
Sbjct 42 CANPTYECSLMRIEGYEYCI-----RHILRDPRSNFRQCSHVYSNGRKCIN 87
> At1g64310
Length=552
Score = 27.7 bits (60), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 18/43 (41%), Gaps = 12/43 (27%)
Query 26 PQMDTCSGLKEQYDRCFNHWYRRHFLRGDMSRNCFHLFADYRL 68
P + CS L Y RC NH + HLFA+ R+
Sbjct 271 PDLVACSSLITGYSRCGNH------------KEALHLFAELRM 301
> CE16248_1
Length=1259
Score = 27.7 bits (60), Expect = 6.0, Method: Composition-based stats.
Identities = 13/41 (31%), Positives = 21/41 (51%), Gaps = 2/41 (4%)
Query 28 MDTCSGLKEQYDRCFNHWYRRHFLRGDMSRNCFHLFADYRL 68
+D +K YD + HWY ++ D S N +H + Y+L
Sbjct 432 LDDLKHMKHNYDTMYKHWYDMYW-DADESANEYH-YPPYKL 470
> YDR031w
Length=117
Score = 27.3 bits (59), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 14/53 (26%), Positives = 24/53 (45%), Gaps = 4/53 (7%)
Query 28 MDTCSGLKEQYDRCFNHWYRRHFLRGDMSRNCFHLFADYRLCISEELKRKGLE 80
M CS ++YD+C R + ++ NC D R C ++K K ++
Sbjct 60 MSECSEPMKKYDQCI----RDNMGTRTINENCLGFLQDLRKCAELQVKNKNIK 108
> Hs4885643
Length=1005
Score = 26.9 bits (58), Expect = 9.1, Method: Composition-based stats.
Identities = 21/73 (28%), Positives = 29/73 (39%), Gaps = 7/73 (9%)
Query 5 SGIAPSAYVDADFWTAAAACAPQMDTCSGLKEQYDRCFNHWYRRHFLRGDMSRNCFHLFA 64
SG A A +D TAA D C ++E Y +C Y G M++ +
Sbjct 891 SGAAVFAMTYSDMQTAA-------DKCEEMEEGYTQCSQFLYGVQEKMGIMNKGVIYALW 943
Query 65 DYRLCISEELKRK 77
DY +EL K
Sbjct 944 DYEPQNDDELPMK 956
Lambda K H
0.326 0.137 0.466
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1165602088
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40