bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0794_orf1
Length=168
Score E
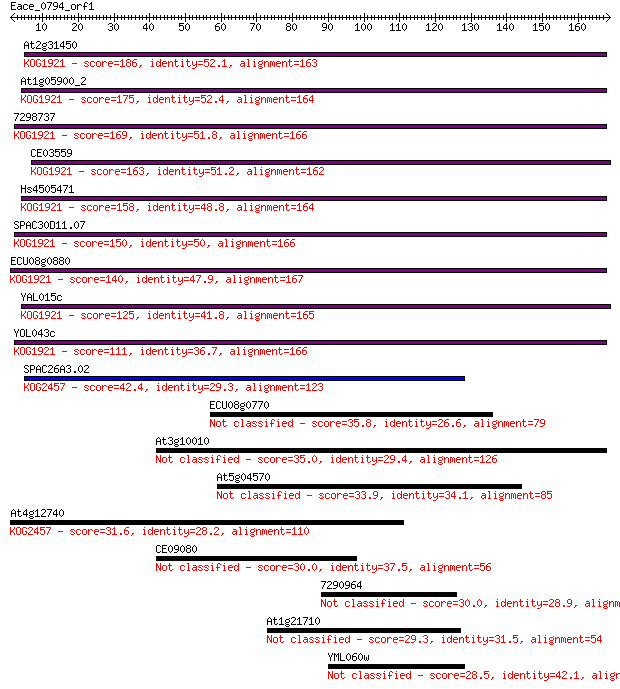
Sequences producing significant alignments: (Bits) Value
At2g31450 186 1e-47
At1g05900_2 175 3e-44
7298737 169 3e-42
CE03559 163 2e-40
Hs4505471 158 6e-39
SPAC30D11.07 150 1e-36
ECU08g0880 140 1e-33
YAL015c 125 4e-29
YOL043c 111 8e-25
SPAC26A3.02 42.4 4e-04
ECU08g0770 35.8 0.042
At3g10010 35.0 0.070
At5g04570 33.9 0.17
At4g12740 31.6 0.84
CE09080 30.0 2.1
7290964 30.0 2.3
At1g21710 29.3 4.0
YML060w 28.5 5.8
> At2g31450
Length=281
Score = 186 bits (473), Expect = 1e-47, Method: Compositional matrix adjust.
Identities = 85/170 (50%), Positives = 119/170 (70%), Gaps = 7/170 (4%)
Query 5 ERRFAVLVAVMLSSQTKDEQTHACVNRLRENGLLNPYAIAACDPEKLRGLLFGVGFHNNK 64
ERRFAVL+ +LSSQTKD+ +A ++RL +NGLL P A+ D ++ L++ VGF+ K
Sbjct 73 ERRFAVLLGALLSSQTKDQVNNAAIHRLHQNGLLTPEAVDKADESTIKELIYPVGFYTRK 132
Query 65 TTFLKEAAEILIKKHKGEVPQTLDDLMQLRGVGRKMANIVMHAAWNSFHGIAVDVHVHRI 124
T++K+ A I + K+ G++P +LDDL+ L G+G KMA++++H AWN GI VD HVHRI
Sbjct 133 ATYMKKIARICLVKYDGDIPSSLDDLLSLPGIGPKMAHLILHIAWNDVQGICVDTHVHRI 192
Query 125 SNRLGWV-------KTKTPQETEIALEDCLPRKHWEDVNLLLVGFGQQIC 167
NRLGWV KT +P+ET +AL+ LP++ W +N LLVGFGQ IC
Sbjct 193 CNRLGWVSRPGTKQKTTSPEETRVALQQWLPKEEWVAINPLLVGFGQMIC 242
> At1g05900_2
Length=307
Score = 175 bits (444), Expect = 3e-44, Method: Compositional matrix adjust.
Identities = 86/187 (45%), Positives = 117/187 (62%), Gaps = 23/187 (12%)
Query 4 KERRFAVLVAVMLSSQTKDEQTHACVNRLRENGLLNPYAIAACDPEKLRGLLFGVGFHNN 63
KERRF VL+ +LSSQTK+ T A V RL +NGLL P AI D ++ L++ VGF+
Sbjct 82 KERRFYVLIGTLLSSQTKEHITGAAVERLHQNGLLTPEAIDKADESTIKELIYPVGFYTR 141
Query 64 KTTFLKEAAEILIKKHKGEVPQTLDDLMQLRGVGRKMANIVMHAAWNSFHGIAVDVHVHR 123
K T +K+ A+I + ++ G++P+TL++L+ L GVG K+A++V+H AWN GI VD HVHR
Sbjct 142 KATNVKKVAKICLMEYDGDIPRTLEELLSLPGVGPKIAHLVLHVAWNDVQGICVDTHVHR 201
Query 124 ISNRLGWV-----------------------KTKTPQETEIALEDCLPRKHWEDVNLLLV 160
I NRLGWV KT +P+ET +AL+ LP+ W +N LLV
Sbjct 202 ICNRLGWVSKPGTKQFAYLLLVTYLYFVLDQKTSSPEETRVALQQWLPKGEWVAINFLLV 261
Query 161 GFGQQIC 167
GFGQ IC
Sbjct 262 GFGQTIC 268
> 7298737
Length=308
Score = 169 bits (428), Expect = 3e-42, Method: Compositional matrix adjust.
Identities = 86/168 (51%), Positives = 109/168 (64%), Gaps = 3/168 (1%)
Query 2 DEKERRFAVLVAVMLSSQTKDEQTHACVNRLRENGLLNPYAIAACDPEKLRGLLFGVGFH 61
D K +RF LVA+MLSSQTKD+ T+ +NRL++ GL P + +L LL V F+
Sbjct 111 DSKTQRFQNLVALMLSSQTKDQTTYEAMNRLKDRGL-TPLKVKEMPVTELENLLHPVSFY 169
Query 62 NNKTTFLKEAAEILIKKHKGEVPQTLDDLMQLRGVGRKMANIVMHAAWNSFHGIAVDVHV 121
NK +LK+ EIL K+ ++P + DL+ L GVG KMA+I M AWN GI VDVHV
Sbjct 170 KNKAKYLKQTVEILTDKYGSDIPDNVKDLVALPGVGPKMAHICMAVAWNKITGIGVDVHV 229
Query 122 HRISNRLGWV--KTKTPQETEIALEDCLPRKHWEDVNLLLVGFGQQIC 167
HR+SNRLGWV TK P++T +ALE LP W +VN L VGFGQ IC
Sbjct 230 HRLSNRLGWVPKPTKEPEQTRVALEKWLPFSLWSEVNHLFVGFGQTIC 277
> CE03559
Length=259
Score = 163 bits (412), Expect = 2e-40, Method: Compositional matrix adjust.
Identities = 83/162 (51%), Positives = 109/162 (67%), Gaps = 1/162 (0%)
Query 7 RFAVLVAVMLSSQTKDEQTHACVNRLRENGLLNPYAIAACDPEKLRGLLFGVGFHNNKTT 66
RF VLVA+MLSSQT+DE A + RL+++GL + P+ L +L VGF+ K
Sbjct 28 RFQVLVALMLSSQTRDEVNAAAMKRLKDHGLSIGKILEFKVPD-LETILCPVGFYKRKAV 86
Query 67 FLKEAAEILIKKHKGEVPQTLDDLMQLRGVGRKMANIVMHAAWNSFHGIAVDVHVHRISN 126
+L++ A+IL G++P +LD L L GVG KMAN+VM AW GIAVD HVHRISN
Sbjct 87 YLQKTAKILKDDFSGDIPDSLDGLCALPGVGPKMANLVMQIAWGECVGIAVDTHVHRISN 146
Query 127 RLGWVKTKTPQETEIALEDCLPRKHWEDVNLLLVGFGQQICK 168
RLGW+KT TP++T+ ALE LP+ W+ +N LLVGFGQ C+
Sbjct 147 RLGWIKTSTPEKTQKALEILLPKSEWQPINHLLVGFGQMQCQ 188
> Hs4505471
Length=312
Score = 158 bits (399), Expect = 6e-39, Method: Compositional matrix adjust.
Identities = 80/166 (48%), Positives = 112/166 (67%), Gaps = 3/166 (1%)
Query 4 KERRFAVLVAVMLSSQTKDEQTHACVNRLRENGLLNPYAIAACDPEKLRGLLFGVGFHNN 63
K RR+ VL+++MLSSQTKD+ T + RLR GL +I D L L++ VGF +
Sbjct 126 KVRRYQVLLSLMLSSQTKDQVTAGAMQRLRARGLTV-DSILQTDDATLGKLIYPVGFWRS 184
Query 64 KTTFLKEAAEILIKKHKGEVPQTLDDLMQLRGVGRKMANIVMHAAWNSFHGIAVDVHVHR 123
K ++K+ + IL + + G++P ++ +L+ L GVG KMA++ M AW + GIAVD HVHR
Sbjct 185 KVKYIKQTSAILQQHYGGDIPASVAELVALPGVGPKMAHLAMAVAWGTVSGIAVDTHVHR 244
Query 124 ISNRLGWVK--TKTPQETEIALEDCLPRKHWEDVNLLLVGFGQQIC 167
I+NRL W K TK+P+ET ALE+ LPR+ W ++N LLVGFGQQ C
Sbjct 245 IANRLRWTKKATKSPEETRAALEEWLPRELWHEINGLLVGFGQQTC 290
> SPAC30D11.07
Length=355
Score = 150 bits (378), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 83/168 (49%), Positives = 104/168 (61%), Gaps = 2/168 (1%)
Query 2 DEKERRFAVLVAVMLSSQTKDEQTHACVNRLREN--GLLNPYAIAACDPEKLRGLLFGVG 59
D K+ RF LVA+MLSSQTKD + L+E G L I D L L+ VG
Sbjct 43 DPKKFRFQTLVALMLSSQTKDIVLGPTMRNLKEKLAGGLCLEDIQNIDEVSLNKLIEKVG 102
Query 60 FHNNKTTFLKEAAEILIKKHKGEVPQTLDDLMQLRGVGRKMANIVMHAAWNSFHGIAVDV 119
FHN KT +LK+ A IL +K +G++P T++DLM L GVG KM + M AWN GI VDV
Sbjct 103 FHNRKTIYLKQMARILSEKFQGDIPDTVEDLMTLPGVGPKMGYLCMSIAWNKTVGIGVDV 162
Query 120 HVHRISNRLGWVKTKTPQETEIALEDCLPRKHWEDVNLLLVGFGQQIC 167
HVHRI N L W TKT ++T AL+ LP++ W ++N LVGFGQ IC
Sbjct 163 HVHRICNLLHWCNTKTEEQTRAALQSWLPKELWFELNHTLVGFGQTIC 210
> ECU08g0880
Length=238
Score = 140 bits (353), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 80/179 (44%), Positives = 110/179 (61%), Gaps = 15/179 (8%)
Query 1 KDEKERRFAVLVAVMLSSQTKDEQTHACVNRLRE------------NGLLNPYAIAACDP 48
+ E+ERRF +LV+++LSSQTKDE T+ + RLR+ G L +A D
Sbjct 42 RTEEERRFHILVSLLLSSQTKDEVTYEAMARLRKLLPESAATDGEARGGLTIERVANSDV 101
Query 49 EKLRGLLFGVGFHNNKTTFLKEAAEILIKKHKGEVPQTLDDLMQLRGVGRKMANIVMHAA 108
+ + + VGFHN K LK+ AEIL + KG +P+ + DL+ L G+G KMA + M A
Sbjct 102 KHINECIKKVGFHNRKAANLKKIAEIL--REKG-LPREMKDLISLPGIGNKMALLYMSHA 158
Query 109 WNSFHGIAVDVHVHRISNRLGWVKTKTPQETEIALEDCLPRKHWEDVNLLLVGFGQQIC 167
N GI+VD HVHRISNR+G V+T+ + T LE +PRK W+ +N +LVGFGQ IC
Sbjct 159 CNRTVGISVDTHVHRISNRIGLVRTRDVESTRRELERVVPRKEWKTINNILVGFGQTIC 217
> YAL015c
Length=399
Score = 125 bits (314), Expect = 4e-29, Method: Compositional matrix adjust.
Identities = 69/176 (39%), Positives = 97/176 (55%), Gaps = 11/176 (6%)
Query 4 KERRFAVLVAVMLSSQTKDEQT--------HACVNRLRENGLLNPYAIAACDPEKLRGLL 55
++ R VL+ VMLSSQTKDE T C++ L + A+ + KL L+
Sbjct 140 RDYRLQVLLGVMLSSQTKDEVTAMAMLNIMRYCIDELHSEEGMTLEAVLQINETKLDELI 199
Query 56 FGVGFHNNKTTFLKEAAEILIKKHKGEVPQTLDDLMQLRGVGRKMANIVMHAAWNSFHGI 115
VGFH K ++ +IL + +VP T+++L+ L GVG KMA + + AW GI
Sbjct 200 HSVGFHTRKAKYILSTCKILQDQFSSDVPATINELLGLPGVGPKMAYLTLQKAWGKIEGI 259
Query 116 AVDVHVHRISNRLGWV---KTKTPQETEIALEDCLPRKHWEDVNLLLVGFGQQICK 168
VDVHV R++ WV K KTP +T L++ LP+ W ++N LLVGFGQ I K
Sbjct 260 CVDVHVDRLTKLWKWVDAQKCKTPDQTRTQLQNWLPKGLWTEINGLLVGFGQIITK 315
> YOL043c
Length=380
Score = 111 bits (277), Expect = 8e-25, Method: Compositional matrix adjust.
Identities = 61/177 (34%), Positives = 89/177 (50%), Gaps = 11/177 (6%)
Query 2 DEKERRFAVLVAVMLSSQTKDEQ--------THACVNRLRENGLLNPYAIAACDPEKLRG 53
D K R L+ MLS+QT+DE+ T C+N L+ + + D L
Sbjct 143 DPKNFRLQFLIGTMLSAQTRDERMAQAALNITEYCLNTLKIAEGITLDGLLKIDEPVLAN 202
Query 54 LLFGVGFHNNKTTFLKEAAEILIKKHKGEVPQTLDDLMQLRGVGRKMANIVMHAAWNSFH 113
L+ V F+ K F+K A++L+ ++P ++ ++ L GVG KM + + W
Sbjct 203 LIRCVSFYTRKANFIKRTAQLLVDNFDSDIPYDIEGILSLPGVGPKMGYLTLQKGWGLIA 262
Query 114 GIAVDVHVHRISNRLGWV---KTKTPQETEIALEDCLPRKHWEDVNLLLVGFGQQIC 167
GI VDVHVHR+ WV K KT + T L+ LP W ++N +LVGFGQ IC
Sbjct 263 GICVDVHVHRLCKMWNWVDPIKCKTAEHTRKELQVWLPHSLWYEINTVLVGFGQLIC 319
> SPAC26A3.02
Length=461
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 36/124 (29%), Positives = 59/124 (47%), Gaps = 3/124 (2%)
Query 5 ERRFAVLVAVMLSSQTKDEQTHACVNRLRENGLLNPYAIAACDPEKLRGLLFGVGFHNNK 64
+R + VLV+ ++ QT+ E + E A ++ L G+GF+ +
Sbjct 60 QRLYEVLVSEIMLQQTRVETVKRYYTKWMETLPTLKSCAEAEYNTQVMPLWSGMGFYT-R 118
Query 65 TTFLKEAAEILIKKHKGEVPQTLDDLMQ-LRGVGRKMANIVMHAAWNSFHGIAVDVHVHR 123
L +A + L K H E+P+T D+ + + GVG A V+ AW GI VD +V R
Sbjct 119 CKRLHQACQHLAKLHPSEIPRTGDEWAKGIPGVGPYTAGAVLSIAWKQPTGI-VDGNVIR 177
Query 124 ISNR 127
+ +R
Sbjct 178 VLSR 181
> ECU08g0770
Length=290
Score = 35.8 bits (81), Expect = 0.042, Method: Compositional matrix adjust.
Identities = 21/81 (25%), Positives = 43/81 (53%), Gaps = 5/81 (6%)
Query 57 GVGFHNNKTTFLKEAAEILIKKHK--GEVPQTLDDLMQLRGVGRKMANIVMHAAWNSFHG 114
G G+ ++ ++ AAE L++ + + D ++ ++GVG K+A+ ++ +
Sbjct 168 GFGY---RSRYICSAAEYLMENYPRLQQASGVRDMMVSIKGVGDKIADCILLIGLGNLSV 224
Query 115 IAVDVHVHRISNRLGWVKTKT 135
+ VD H+ R S +L V+ KT
Sbjct 225 VPVDTHIFRHSRKLFGVRGKT 245
> At3g10010
Length=1309
Score = 35.0 bits (79), Expect = 0.070, Method: Compositional matrix adjust.
Identities = 37/138 (26%), Positives = 62/138 (44%), Gaps = 21/138 (15%)
Query 42 AIAACDPEKLRGLLFGVGFHNNKTTFLKEAAEILIKKHKG-------EVP--QTLDDLMQ 92
A+ D K+ ++ G +N +K L+KKH +VP + + L+
Sbjct 819 ALRCTDVHKIANIIIKRGMNNMLAERIKAFLNRLVKKHGSIDLEWLRDVPPDKAKEYLLS 878
Query 93 LRGVGRKMANIVMHAAWNSFHGIA--VDVHVHRISNRLGWVKTKT-PQETEIALEDCLPR 149
+ G+G K V S H IA VD +V RI+ RLGWV + P E ++ L +
Sbjct 879 INGLGLKSVECVRLL---SLHQIAFPVDTNVGRIAVRLGWVPLQPLPDELQMHLLELY-- 933
Query 150 KHWEDVNLLLVGFGQQIC 167
+++ ++ FG+ C
Sbjct 934 ----ELHYHMITFGKVFC 947
> At5g04570
Length=555
Score = 33.9 bits (76), Expect = 0.17, Method: Composition-based stats.
Identities = 29/97 (29%), Positives = 47/97 (48%), Gaps = 15/97 (15%)
Query 59 GFHNNKTTFLKEAAEILIKKHKG-------EVP--QTLDDLMQLRGVGRKMANIVMHAAW 109
G +N +K+ E ++K H G E P + D L+ +RG+G K V
Sbjct 459 GMNNMLAVRIKDFLERIVKDHGGIDLEWLRESPPDKAKDYLLSIRGLGLKSVECVRLL-- 516
Query 110 NSFHGIA--VDVHVHRISNRLGWVKTKT-PQETEIAL 143
+ H +A VD +V RI+ R+GWV + P+ ++ L
Sbjct 517 -TLHNLAFPVDTNVGRIAVRMGWVPLQPLPESLQLHL 552
> At4g12740
Length=608
Score = 31.6 bits (70), Expect = 0.84, Method: Compositional matrix adjust.
Identities = 31/129 (24%), Positives = 59/129 (45%), Gaps = 21/129 (16%)
Query 1 KDEKERR-FAVLVAVMLSSQTKDE----------QTHACVNRLRENGLLNPYAIAACDPE 49
+ EKERR + V V+ ++ QT+ + Q + L + L N + +
Sbjct 154 ESEKERRAYEVWVSEIMLQQTRVQTVMKYYKRWMQKWPTIYDLGQASLENLIVSRSRELS 213
Query 50 KLRG--------LLFGVGFHNNKTTFLKEAAEILIKKHKGEVPQTLDDLMQLRGVGRKMA 101
LRG + G+G++ + FL E A++++ +G P LM+++G+G+ A
Sbjct 214 FLRGNEKKEVNEMWAGLGYYR-RARFLLEGAKMVVAGTEG-FPNQASSLMKVKGIGQYTA 271
Query 102 NIVMHAAWN 110
+ A+N
Sbjct 272 GAIASIAFN 280
> CE09080
Length=508
Score = 30.0 bits (66), Expect = 2.1, Method: Composition-based stats.
Identities = 21/59 (35%), Positives = 29/59 (49%), Gaps = 6/59 (10%)
Query 42 AIAACDPEKLRGL---LFGVGFHNNKTTFLKEAAEILIKKHKGEVPQTLDDLMQLRGVG 97
A DP +RGL LFG+ N T L E+L+ K + +DL+QLR +G
Sbjct 113 AAQTSDP-NIRGLAAELFGLRISNQNLTMLLNTLEVLLSATK--IKNVSNDLLQLRTLG 168
> 7290964
Length=327
Score = 30.0 bits (66), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 11/38 (28%), Positives = 23/38 (60%), Gaps = 0/38 (0%)
Query 88 DDLMQLRGVGRKMANIVMHAAWNSFHGIAVDVHVHRIS 125
++L L G+G K+A+ + + + VD+H++RI+
Sbjct 223 EELTLLPGIGYKVADCICLMSMGHLESVPVDIHIYRIA 260
> At1g21710
Length=365
Score = 29.3 bits (64), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 17/54 (31%), Positives = 27/54 (50%), Gaps = 0/54 (0%)
Query 73 EILIKKHKGEVPQTLDDLMQLRGVGRKMANIVMHAAWNSFHGIAVDVHVHRISN 126
E L+ K E+ + + L L GVG K+A + + + I VD HV +I+
Sbjct 235 EWLLSLRKVELQEAVAALCTLPGVGPKVAACIALFSLDQHSAIPVDTHVWQIAT 288
> YML060w
Length=376
Score = 28.5 bits (62), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 16/38 (42%), Positives = 21/38 (55%), Gaps = 0/38 (0%)
Query 90 LMQLRGVGRKMANIVMHAAWNSFHGIAVDVHVHRISNR 127
LM GVG K+A+ V + + VDVHV RI+ R
Sbjct 232 LMSYNGVGPKVADCVCLMGLHMDGIVPVDVHVSRIAKR 269
Lambda K H
0.321 0.136 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2418402922
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40