bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
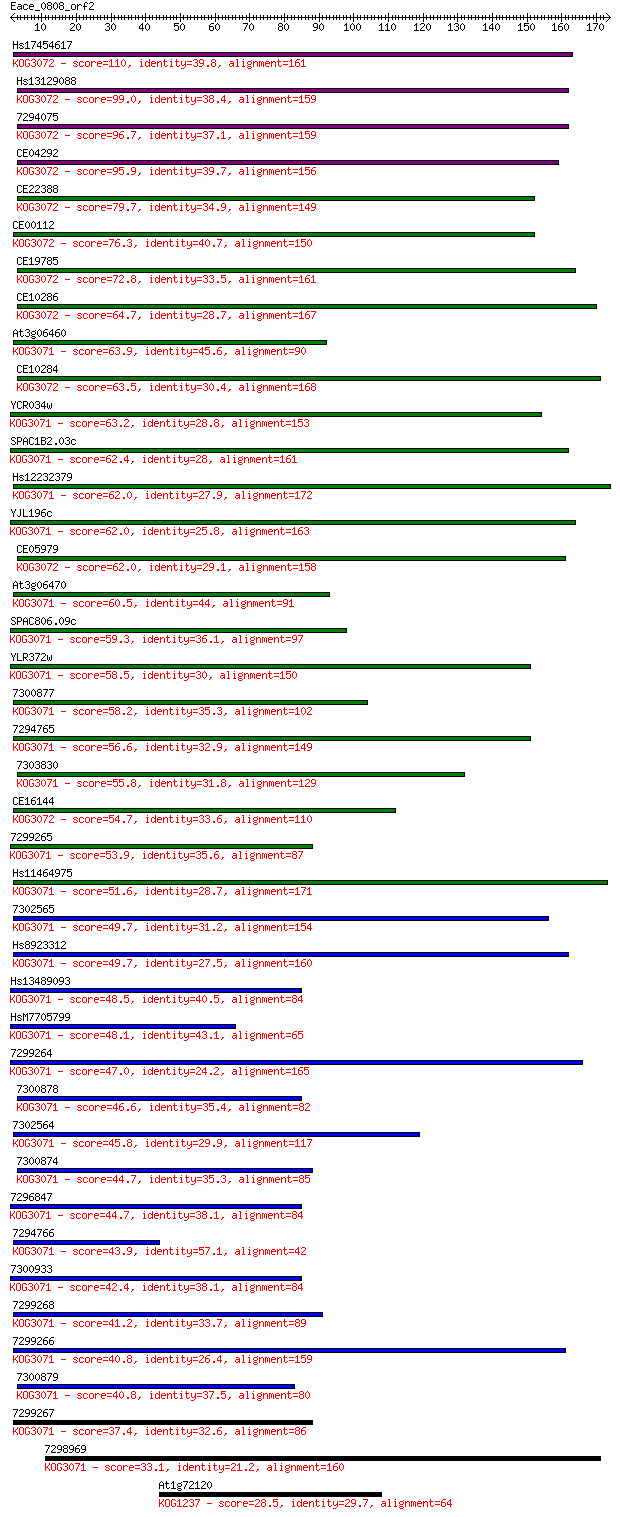
Query= Eace_0808_orf2
Length=173
Score E
Sequences producing significant alignments: (Bits) Value
Hs17454617 110 2e-24
Hs13129088 99.0 4e-21
7294075 96.7 2e-20
CE04292 95.9 4e-20
CE22388 79.7 3e-15
CE00112 76.3 3e-14
CE19785 72.8 3e-13
CE10286 64.7 8e-11
At3g06460 63.9 2e-10
CE10284 63.5 2e-10
YCR034w 63.2 2e-10
SPAC1B2.03c 62.4 4e-10
Hs12232379 62.0 5e-10
YJL196c 62.0 5e-10
CE05979 62.0 6e-10
At3g06470 60.5 2e-09
SPAC806.09c 59.3 4e-09
YLR372w 58.5 7e-09
7300877 58.2 8e-09
7294765 56.6 2e-08
7303830 55.8 4e-08
CE16144 54.7 9e-08
7299265 53.9 2e-07
Hs11464975 51.6 7e-07
7302565 49.7 3e-06
Hs8923312 49.7 3e-06
Hs13489093 48.5 7e-06
HsM7705799 48.1 8e-06
7299264 47.0 2e-05
7300878 46.6 2e-05
7302564 45.8 4e-05
7300874 44.7 1e-04
7296847 44.7 1e-04
7294766 43.9 2e-04
7300933 42.4 5e-04
7299268 41.2 0.001
7299266 40.8 0.001
7300879 40.8 0.001
7299267 37.4 0.016
7298969 33.1 0.32
At1g72120 28.5 7.7
> Hs17454617
Length=270
Score = 110 bits (275), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 64/162 (39%), Positives = 97/162 (59%), Gaps = 18/162 (11%)
Query 2 LELIDTAFIILRKKRLSFLHYYHHATVLLYTWDAYTCEQPAGIYFVAMNYTVHAAMYLYF 61
+EL DTAFIILRK+ L F+H+YHH+TVL+YT Y + PAG +FV MN+ VHA MY Y+
Sbjct 126 IELGDTAFIILRKRPLIFIHWYHHSTVLVYTSFGYKNKVPAGGWFVTMNFGVHAIMYTYY 185
Query 62 FI-AAVLERPLSWGIFVTVAQISQMFIGVGVTFVSMYIACTAPYKALWHRQQLDDALSYG 120
+ AA ++ P + +T QI QMF+G V+ ++ +W + Q G
Sbjct 186 TLKAANVKPPKMLPMLITSLQILQMFVGAIVSILTY----------IWRQDQ-------G 228
Query 121 HYITLKNLYWACLMYSTYFYLFAEYFVKRYLRKPQNNKHQQH 162
+ T+++L+W+ ++Y TYF LFA +F + Y+R K +
Sbjct 229 CHTTMEHLFWSFILYMTYFILFAHFFCQTYIRPKVKAKTKSQ 270
> Hs13129088
Length=265
Score = 99.0 bits (245), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 61/160 (38%), Positives = 91/160 (56%), Gaps = 18/160 (11%)
Query 3 ELIDTAFIILRKKRLSFLHYYHHATVLLYTWDAYTCEQPAGIYFVAMNYTVHAAMYLYFF 62
EL DT FIILRK++L FLH+YHH TVLLY+W +Y G +F+ MNY VHA MY Y+
Sbjct 123 ELGDTIFIILRKQKLIFLHWYHHITVLLYSWYSYKDMVAGGGWFMTMNYGVHAVMYSYYA 182
Query 63 I-AAVLERPLSWGIFVTVAQISQMFIGVGVTFVSMYIACTAPYKALWHRQQLDDALSYGH 121
+ AA + +F+T++QI+QM +G V ++ + C W Q D S+
Sbjct 183 LRAAGFRVSRKFAMFITLSQITQMLMGCVVNYL---VFC-------W--MQHDQCHSH-- 228
Query 122 YITLKNLYWACLMYSTYFYLFAEYFVKRYLRKPQNNKHQQ 161
+N++W+ LMY +Y LF +F + Y+ K + +
Sbjct 229 ---FQNIFWSSLMYLSYLVLFCHFFFEAYIGKMRKTTKAE 265
> 7294075
Length=313
Score = 96.7 bits (239), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 59/160 (36%), Positives = 88/160 (55%), Gaps = 16/160 (10%)
Query 3 ELIDTAFIILRKKRLSFLHYYHHATVLLYTWDAYTCEQPAGIYFVAMNYTVHAAMYLYFF 62
EL DT FI+LRK+ L FLH+YHH TVL+Y+W +YT + +F+ MNY VH+ MY Y+
Sbjct 126 ELGDTIFIVLRKQPLIFLHWYHHITVLIYSWFSYTEYTSSARWFIVMNYCVHSVMYSYYA 185
Query 63 I-AAVLERPLSWGIFVTVAQISQMFIGVGVTFVSMYIACTAPYKALWHRQQLDDALSYGH 121
+ AA P + +T Q++QM IG + +W L +
Sbjct 186 LKAARFNPPRFISMIITSLQLAQMIIGCAIN--------------VWANGFLKTHGTSSC 231
Query 122 YITLKNLYWACLMYSTYFYLFAEYFVKRYLRKPQNNKHQQ 161
+I+ +N+ + MYS+YF LFA +F K YL P +K ++
Sbjct 232 HISQRNINLSIAMYSSYFVLFARFFYKAYL-APGGHKSRR 270
> CE04292
Length=435
Score = 95.9 bits (237), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 62/158 (39%), Positives = 88/158 (55%), Gaps = 21/158 (13%)
Query 3 ELIDTAFIILRKKRLSFLHYYHHATVLLYTWDAYTCEQPAGIYFVAMNYTVHAAMYLYFF 62
ELIDT FI+LRK+ L FLH+YHH TV++YTW AY +G +F+ MNY VHA MY Y+
Sbjct 168 ELIDTIFIVLRKRPLIFLHWYHHVTVMIYTWHAYKDHTASGRWFIWMNYGVHALMYSYYA 227
Query 63 IAAVLER-PLSWGIFVTVAQISQMFIGVGVTFVSMYIACTAPYKALWHRQQLDD-ALSYG 120
+ ++ R P + VT Q++QM +GV + I + Y +Q D+ L +G
Sbjct 228 LRSLKFRLPKQMAMVVTTLQLAQMVMGVIIGVTVYRIKSSGEY----CQQTWDNLGLCFG 283
Query 121 HYITLKNLYWACLMYSTYFYLFAEYFVKRYLRKPQNNK 158
+Y TYF LFA +F Y++K NN+
Sbjct 284 -------------VYFTYFLLFANFFYHAYVKK--NNR 306
> CE22388
Length=286
Score = 79.7 bits (195), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 52/151 (34%), Positives = 80/151 (52%), Gaps = 19/151 (12%)
Query 3 ELIDTAFIILRKKRLSFLHYYHHATVLLYTWDAYTCEQPAGIYFVAMNYTVHAAMYLYFF 62
E DT F++LRK+ + FLH+YHHA VL+ +W A G +F+ MNY VH+ MY Y+
Sbjct 141 EFGDTMFLVLRKRPVIFLHWYHHAVVLILSWHAAIELTAPGRWFIFMNYLVHSIMYTYYA 200
Query 63 IAAVLER-PLSWGIFVTVAQISQMFIGVGVTFVSMYIACTAPYKALWHRQQLDD-ALSYG 120
I ++ R P + VT Q QM IGV ++ + +Y+ +Q D+ ALS+G
Sbjct 201 ITSIGYRLPKIVSMTVTFLQTLQMLIGVSISCIVLYLKLNGEM----CQQSYDNLALSFG 256
Query 121 HYITLKNLYWACLMYSTYFYLFAEYFVKRYL 151
+Y+++ LF+ +F YL
Sbjct 257 -------------IYASFLVLFSSFFNNAYL 274
> CE00112
Length=291
Score = 76.3 bits (186), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 61/151 (40%), Positives = 85/151 (56%), Gaps = 16/151 (10%)
Query 2 LELIDTAFIILRKKRLSFLHYYHHATVLLYTWDAYTCEQPAGIYFVAMNYTVHAAMYLYF 61
+EL DT FIILRK+ L FLHYYHHA VL+YT + AG +++ MNY H+ MY Y+
Sbjct 143 VELGDTMFIILRKRPLIFLHYYHHAAVLIYTVHSGAEHTAAGRFYILMNYFAHSLMYTYY 202
Query 62 FIAAVLERPLSW-GIFVTVAQISQMFIGVGVTFVSMYIACTAPYKALWHRQQLDDALSYG 120
++A+ R W + VT Q +QM GVG+T+ M YK QQ
Sbjct 203 TVSAMGYRLPKWVSMTVTTVQTTQMLAGVGITW--MVYKVKTEYKL--PCQQ-------- 250
Query 121 HYITLKNLYWACLMYSTYFYLFAEYFVKRYL 151
++ NLY A ++Y T+ LF ++FVK Y+
Sbjct 251 ---SVANLYLAFVIYVTFAILFIQFFVKAYI 278
> CE19785
Length=274
Score = 72.8 bits (177), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 54/163 (33%), Positives = 80/163 (49%), Gaps = 17/163 (10%)
Query 3 ELIDTAFIILRKKRLSFLHYYHHATVLLYTWDAYTCEQPAGIYFVAMNYTVHAAMYLYFF 62
EL DT F++LRKK + F+H+YHHA +Y Y+ Q + +A+N VH MY YF
Sbjct 127 ELGDTMFLVLRKKPVIFMHWYHHALTFVYAVVTYSEHQAWARWSLALNLAVHTVMYFYFA 186
Query 63 IAAV-LERPLSWGIFVTVAQISQMFIGVGVTFVSMYIACTAPYKALWHRQQLDDALSY-G 120
+ A+ ++ P F+T QI Q I S YI H + A S G
Sbjct 187 VRALNIQTPRPVAKFITTIQIVQFVI-------SCYI--------FGHLVFIKSADSVPG 231
Query 121 HYITLKNLYWACLMYSTYFYLFAEYFVKRYLRKPQNNKHQQHQ 163
++ L LMY +Y +LFA++F K Y++K K + +
Sbjct 232 CAVSWNVLSIGGLMYISYLFLFAKFFYKAYIQKRSPTKTSKQE 274
> CE10286
Length=274
Score = 64.7 bits (156), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 48/169 (28%), Positives = 76/169 (44%), Gaps = 19/169 (11%)
Query 3 ELIDTAFIILRKKRLSFLHYYHHATVLLYTWDAYTCEQPAGIYFVAMNYTVHAAMYLYFF 62
E DT FIILRKK L FLH+YHH + Y + ++ + MN++VH+ MY Y+
Sbjct 123 EFGDTLFIILRKKPLMFLHWYHHVLTMNYAFMSFEANLGFNTWITWMNFSVHSIMYGYYM 182
Query 63 IAAVLERPLSW-GIFVTVAQISQMFIGVGVTFVSMYIACTAPYKALWHRQQLDDALSYGH 121
+ + + +W +T QI Q I + F Y+A T Q +D Y
Sbjct 183 LRSFGVKVPAWIAKNITTMQILQFVITHFILFHVGYLAVTG--------QSVDSTPGY-- 232
Query 122 YITLKNLYWAC-LMYSTYFYLFAEYFVKRYLRKPQNNKHQQHQLQHPKE 169
YW C LM +Y LF ++ + Y++ + + + + E
Sbjct 233 -------YWFCLLMEISYVVLFGNFYYQSYIKGGGKKFNAEKKTEKKIE 274
> At3g06460
Length=298
Score = 63.9 bits (154), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 41/95 (43%), Positives = 54/95 (56%), Gaps = 9/95 (9%)
Query 2 LELIDTAFIILRK--KRLSFLHYYHHATVLLYTWDAYTCEQ---PAGIYFVAMNYTVHAA 56
LE +DT IIL K +RLSFLH YHHATV++ + Q P G+ +N TVH
Sbjct 130 LEFVDTLLIILNKSIQRLSFLHVYHHATVVILCYLWLRTRQSMFPVGL---VLNSTVHVI 186
Query 57 MYLYFFIAAVLERPLSWGIFVTVAQISQMFIGVGV 91
MY Y+F+ A+ RP W VT Q+ Q G+G+
Sbjct 187 MYGYYFLCAIGSRP-KWKKLVTNFQMVQFAFGMGL 220
> CE10284
Length=286
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 51/172 (29%), Positives = 88/172 (51%), Gaps = 18/172 (10%)
Query 3 ELIDTAFIILRKKRLSFLHYYHHATVLLYTWDAYTCEQPAGIYFVAMNYTVHAAMYLYFF 62
EL+DT FI+LRK+ L F+H+YHHA Y Y + ++ V MNY +HA MY Y+
Sbjct 129 ELLDTVFIVLRKRPLIFMHWYHHALTGYYALVCYHEDAVHMVWVVWMNYIIHAFMYGYYL 188
Query 63 IAAV-LERPLSWGIFVTVAQISQMFIGVGVTFVSMYIACTAPYKALWHRQQLDDALSYGH 121
+ ++ + P S +T +Q+ Q + + F ++++ YK H + + L+Y
Sbjct 189 LKSLKVPIPPSVAQAITTSQMVQFAVAI---FAQVHVS----YK---HYVEGVEGLAY-- 236
Query 122 YITLKNLYWACLMYSTYFYLFAEYFVKRYLR---KPQNNKHQQHQLQHPKEH 170
+ + M +TYFYL+ +++ + YL+ K N Q + Q K +
Sbjct 237 --SFRGTAIGFFMLTTYFYLWIQFYKEHYLKNGGKKYNLAKDQAKTQTKKAN 286
> YCR034w
Length=347
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 44/153 (28%), Positives = 72/153 (47%), Gaps = 8/153 (5%)
Query 1 FLELIDTAFIILRKKRLSFLHYYHHATVLLYTWDAYTCEQPAGIYFVAMNYTVHAAMYLY 60
F+E IDT F++L+ K+L+FLH YHH L + +++N VH MY Y
Sbjct 158 FIEFIDTFFLVLKHKKLTFLHTYHHGATALLCYTQLMGTTSISWVPISLNLGVHVVMYWY 217
Query 61 FFIAAVLERPLSWGIFVTVAQISQMFIGVGVTFVSMYIACTAPYKALWHRQQLDDALSYG 120
+F+AA R + W +VT QI Q + +G + ++Y Y + D +
Sbjct 218 YFLAARGIR-VWWKEWVTRFQIIQFVLDIGFIYFAVYQKAVHLYFPIL--PHCGDCVG-- 272
Query 121 HYITLKNLYWACLMYSTYFYLFAEYFVKRYLRK 153
+ + C + S+Y LF +++ Y RK
Sbjct 273 ---STTATFAGCAIISSYLVLFISFYINVYKRK 302
> SPAC1B2.03c
Length=334
Score = 62.4 bits (150), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 45/161 (27%), Positives = 75/161 (46%), Gaps = 8/161 (4%)
Query 1 FLELIDTAFIILRKKRLSFLHYYHHATVLLYTWDAYTCEQPAGIYFVAMNYTVHAAMYLY 60
+LEL+DT F+ L+KK L+FLH YHH L + + +N VH MY Y
Sbjct 145 YLELMDTVFLFLKKKPLAFLHCYHHGITALLCFTQLLGRTSVQWGVIGLNLYVHVIMYSY 204
Query 61 FFIAAVLERPLSWGIFVTVAQISQMFIGVGVTFVSMYIACTAPYKALWHRQQLDDALSYG 120
+F+AA R + W +VT QI Q + + + + Y Y W D + S
Sbjct 205 YFLAAC-GRRVWWKQWVTRVQIIQFVLDLILCYFGTYSHIAFRYFP-WLPHVGDCSGS-- 260
Query 121 HYITLKNLYWACLMYSTYFYLFAEYFVKRYLRKPQNNKHQQ 161
L ++ C + S+Y +LF +++ Y+++ ++
Sbjct 261 ----LFAAFFGCGVLSSYLFLFIGFYINTYIKRGAKKNQRK 297
> Hs12232379
Length=314
Score = 62.0 bits (149), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 48/181 (26%), Positives = 85/181 (46%), Gaps = 31/181 (17%)
Query 2 LELIDTAFIILRKK--RLSFLHYYHHATVLLYTWDAYTCEQPAGIYFVA-MNYTVHAAMY 58
+E +DT F ILRKK ++SFLH YHH T+ W +F A +N +H MY
Sbjct 137 VEYLDTVFFILRKKNNQVSFLHVYHHCTMFTLWWIGIKWVAGGQAFFGAQLNSFIHVIMY 196
Query 59 LYFFIAAV---LERPLSWGIFVTVAQISQMFIGVGVTFVSMYIACTAPYKALWHRQQLDD 115
Y+ + A +++ L W ++T+ Q+ Q + +G T +S+Y C P
Sbjct 197 SYYGLTAFGPWIQKYLWWKRYLTMLQLIQFHVTIGHTALSLYTDCPFP------------ 244
Query 116 ALSYGHYITLKNLYWACLMYS-TYFYLFAEYFVKRYL--RKPQNNKHQQHQLQHPKEHQH 172
K ++WA + Y+ ++ +LF ++++ Y +KP+ K + + +
Sbjct 245 ----------KWMHWALIAYAISFIFLFLNFYIRTYKEPKKPKAGKTAMNGISANGVSKS 294
Query 173 E 173
E
Sbjct 295 E 295
> YJL196c
Length=310
Score = 62.0 bits (149), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 42/163 (25%), Positives = 77/163 (47%), Gaps = 6/163 (3%)
Query 1 FLELIDTAFIILRKKRLSFLHYYHHATVLLYTWDAYTCEQPAGIYFVAMNYTVHAAMYLY 60
F+E DT ++L+ ++L+FLH YHH L ++ V +N VH MY Y
Sbjct 152 FVEFADTVLMVLKHRKLTFLHTYHHGATALLCYNQLVGYTAVTWVPVTLNLAVHVLMYWY 211
Query 61 FFIAAVLERPLSWGIFVTVAQISQMFIGVGVTFVSMYIACTAPYKALWHRQQLDDALSYG 120
+F++A R + W +VT QI Q + + V + +Y A Y Q +D L
Sbjct 212 YFLSASGIR-VWWKAWVTRLQIVQFMLDLIVVYYVLYQKIVAAYFKNACTPQCEDCLG-- 268
Query 121 HYITLKNLYWACLMYSTYFYLFAEYFVKRYLRKPQNNKHQQHQ 163
++ + + ++Y +LF ++++ Y R + K + ++
Sbjct 269 ---SMTAIAAGAAILTSYLFLFISFYIEVYKRGSASGKKKINK 308
> CE05979
Length=288
Score = 62.0 bits (149), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 46/159 (28%), Positives = 73/159 (45%), Gaps = 16/159 (10%)
Query 3 ELIDTAFIILRKKRLSFLHYYHHATVLLYTWDAYTCEQPAGIYFVAMNYTVHAAMYLYFF 62
EL+DT F++LRK+ L FLH+YHH ++Y W ++ Y + +N+ VHA MY Y+F
Sbjct 137 ELVDTIFLVLRKRPLMFLHWYHHILTMIYAWYSHPLTPGFNRYGIYLNFVVHAFMYSYYF 196
Query 63 IAAVLER-PLSWGIFVTVAQISQMFIGVGVTFVSMYIACTAPYKALWHRQQLDDALSYGH 121
+ ++ R P +T QI Q I V L H L +
Sbjct 197 LRSMKIRVPGFIAQAITSLQIVQFIISCAV---------------LAHLGYLMHFTNANC 241
Query 122 YITLKNLYWACLMYSTYFYLFAEYFVKRYLRKPQNNKHQ 160
A M +TY LF +F++ Y+ + +K++
Sbjct 242 DFEPSVFKLAVFMDTTYLALFVNFFLQSYVLRGGKDKYK 280
> At3g06470
Length=278
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 40/93 (43%), Positives = 49/93 (52%), Gaps = 3/93 (3%)
Query 2 LELIDTAFIILRK--KRLSFLHYYHHATVLLYTWDAYTCEQPAGIYFVAMNYTVHAAMYL 59
LE DT IIL K +RLSFLH YHHATV++ + Q + N TVH MY
Sbjct 133 LEFGDTILIILGKSIQRLSFLHVYHHATVVVMCYLWLRTRQSMFPIALVTNSTVHVIMYG 192
Query 60 YFFIAAVLERPLSWGIFVTVAQISQMFIGVGVT 92
Y+F+ AV RP W VT QI Q G++
Sbjct 193 YYFLCAVGSRP-KWKRLVTDCQIVQFVFSFGLS 224
> SPAC806.09c
Length=328
Score = 59.3 bits (142), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 35/97 (36%), Positives = 50/97 (51%), Gaps = 1/97 (1%)
Query 1 FLELIDTAFIILRKKRLSFLHYYHHATVLLYTWDAYTCEQPAGIYFVAMNYTVHAAMYLY 60
FLEL DT F++LRKK L FLH YHH + + + +N VH MY Y
Sbjct 125 FLELTDTFFLVLRKKPLQFLHCYHHGATAVLVYTQIVGRTSISWLIIEINLLVHVTMYYY 184
Query 61 FFIAAVLERPLSWGIFVTVAQISQMFIGVGVTFVSMY 97
+++ A R + W +VT QI Q F +G + ++Y
Sbjct 185 YYLVAKGIR-VPWKKWVTRFQIVQFFADLGFIYFAVY 220
> YLR372w
Length=345
Score = 58.5 bits (140), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 45/157 (28%), Positives = 73/157 (46%), Gaps = 21/157 (13%)
Query 1 FLELIDTAFIILRKKRLSFLHYYHHATVLLYTWDAYTCEQPAGIYFVAMNYTVHAAMYLY 60
F+ELIDT F++LR+K+L FLH YHH L + + +N VH MY Y
Sbjct 165 FVELIDTVFLVLRRKKLLFLHTYHHGATALLCYTQLIGRTSVEWVVILLNLGVHVIMYWY 224
Query 61 FFIAAVLERPLSWGIFVTVAQISQMFIGVGVTFVSM-------YIACTAPYKALWHRQQL 113
+F+++ R + W +VT QI Q I + + + Y+ P K + Q
Sbjct 225 YFLSSCGIR-VWWKQWVTRFQIIQFLIDLVFVYFATYTFYAHKYLDGILPNKGTCYGTQ- 282
Query 114 DDALSYGHYITLKNLYWACLMYSTYFYLFAEYFVKRY 150
A +YG+ L+ ++Y LF ++++ Y
Sbjct 283 -AAAAYGY-----------LILTSYLLLFISFYIQSY 307
> 7300877
Length=322
Score = 58.2 bits (139), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 36/109 (33%), Positives = 57/109 (52%), Gaps = 8/109 (7%)
Query 2 LELIDTAFIILRKK--RLSFLHYYHHATVLLYTWDAYTCEQPA--GIYFVAMNYTVHAAM 57
++L+DT F +LRKK ++SFLH YHH +L++W Y P G+ +N VH M
Sbjct 124 IDLLDTTFFVLRKKDNQVSFLHVYHHTITVLFSW-GYLKYAPGEQGVIIGILNSGVHIIM 182
Query 58 YLYFFIAAVL---ERPLSWGIFVTVAQISQMFIGVGVTFVSMYIACTAP 103
Y Y+ +AA+ ++ L W ++T Q+ Q + +G C P
Sbjct 183 YFYYMVAAMGPQYQKYLWWKKYMTSIQLIQFVLILGYMLTVGAKGCNMP 231
> 7294765
Length=442
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 49/156 (31%), Positives = 77/156 (49%), Gaps = 29/156 (18%)
Query 2 LELIDTAFIILRKK--RLSFLHYYHHATVLLYTWDAYTCEQPAGIYFVAM-NYTVHAAMY 58
LE DTAF ILR+K +LSFLH YHH+T+ ++ W Y AM N VH MY
Sbjct 299 LEFADTAFFILRQKWSQLSFLHVYHHSTMFVFCWILIKWMPTGSTYVPAMINSFVHIIMY 358
Query 59 LYFFIAAV---LERPLSWGIFVTVAQISQMFIGVGVTFVSMYIACTAPYKALWHRQQLDD 115
Y+ ++ + ++R L W ++T Q+ Q I + F W Q L
Sbjct 359 GYYALSVLGPRVQRFLWWKRYLTGLQLVQFTI---IFF--------------WASQMLVR 401
Query 116 ALSYGHYITLKNLYWACLMYS-TYFYLFAEYFVKRY 150
YG +ITL + +YS + ++F ++++++Y
Sbjct 402 GCEYGTWITL-----SMAIYSLPFLFMFGKFYMQKY 432
> 7303830
Length=1280
Score = 55.8 bits (133), Expect = 4e-08, Method: Composition-based stats.
Identities = 41/138 (29%), Positives = 65/138 (47%), Gaps = 10/138 (7%)
Query 3 ELIDTAFIILRKKR--LSFLHYYHH--ATVLLYTWDAYTCEQPAGIYFVAMNYTVHAAMY 58
ELI+T +LRKK+ +S LH +HH L+Y + A + V +N VH MY
Sbjct 115 ELIETVIFVLRKKQNQVSKLHIFHHFSTVTLVYALINFNENGSAAYFCVFLNSIVHVIMY 174
Query 59 LYFFIAAVLERPLSWGIF-----VTVAQISQMFIGVGVTFVSMYIACTAPYKALWHRQQL 113
Y+F+AAV ++ L + +TV Q++Q F+ + + C P L + +
Sbjct 175 SYYFVAAVADKTLVQALTPVKKCITVIQMTQ-FVLILTQVAFQLVLCGMPPLVLLYFTTV 233
Query 114 DDALSYGHYITLKNLYWA 131
+ YG Y + Y A
Sbjct 234 ILGMFYGFYDFYNSAYQA 251
> CE16144
Length=281
Score = 54.7 bits (130), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 37/111 (33%), Positives = 55/111 (49%), Gaps = 9/111 (8%)
Query 2 LELIDTAFIILRKKRLSFLHYYHHATVLLYTWDAYTCEQPAGIYFVAMNYTVHAAMYLYF 61
+E +DT F++LRKK L FLH+YHH ++ Y V +N VHA MY Y+
Sbjct 158 VEFVDTFFLVLRKKPLIFLHWYHHMATFVFFCSNYPTPSSQSRVGVIVNLFVHAFMYPYY 217
Query 62 FIAAV-LERPLSWGIFVTVAQISQMFIGVGVTFVSMYIACTAPYKALWHRQ 111
F ++ ++ P + VTV Q++Q F+ CT Y +L Q
Sbjct 218 FTRSMNIKVPAKISMAVTVLQLTQ--------FMCFIYGCTLMYYSLATNQ 260
> 7299265
Length=257
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 31/94 (32%), Positives = 56/94 (59%), Gaps = 7/94 (7%)
Query 1 FLELIDTAFIILRKK--RLSFLHYYHHATVLLYTWDAYTCEQPAGIYF--VAMNYTVHAA 56
F++L++T F +LRKK ++SFLH +HH + + + Y G+ F +N VH
Sbjct 108 FVDLVETIFFVLRKKDRQISFLHVFHHFAMAFFGYLYYCFHGYGGVAFPQCLLNTAVHVI 167
Query 57 MYLYFFIAAV---LERPLSWGIFVTVAQISQMFI 87
MY Y++++++ ++R L W ++T+AQ+ Q I
Sbjct 168 MYAYYYLSSISKEVQRSLWWKKYITIAQLVQFAI 201
> Hs11464975
Length=299
Score = 51.6 bits (122), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 49/176 (27%), Positives = 78/176 (44%), Gaps = 31/176 (17%)
Query 2 LELIDTAFIILRKK--RLSFLHYYHHATVLLYTWDAYTCEQPAGIYFVA-MNYTVHAAMY 58
+E +DT F ILRK +++ LH YHHA++L W YF A +N +H MY
Sbjct 122 IEFMDTFFFILRKNNHQITVLHVYHHASMLNIWWFVMNWVPCGHSYFGATLNSFIHVLMY 181
Query 59 LYFFIAAVLE-RP-LSWGIFVTVAQISQMFIGVGVTFVSMYIACTAPYKALWHRQQLDDA 116
Y+ +++V RP L W ++T Q+ Q + + T + CT P L+
Sbjct 182 SYYGLSSVPSMRPYLWWKKYITQGQLLQFVLTIIQTSCGVIWPCTFPLGWLY-------- 233
Query 117 LSYGHYITLKNLYWACLMYSTYFYLFAEYFVKRYLRKPQNNKHQQHQLQHPKEHQH 172
G+ I+L LF ++++ Y N K + H K+HQ+
Sbjct 234 FQIGYMISL-------------IALFTNFYIQTY-----NKKGASRRKDHLKDHQN 271
> 7302565
Length=221
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 48/168 (28%), Positives = 78/168 (46%), Gaps = 40/168 (23%)
Query 2 LELIDTAFIILRK--KRLSFLHYYHHATVLLYTWDAYTCEQPAGIYFVA---MNYTVHAA 56
L+L+DT F +LRK K+++FLH YHH + ++ A T G + A +N VH
Sbjct 72 LDLMDTVFFVLRKSYKQITFLHIYHHVFMSFGSY-ALTRYYGTGGHVNAVGLLNSLVHTV 130
Query 57 MYLYFFIAAVLERP-----LSWGIFVTVAQISQMFI----GVGVTFVSMYIACTAPYKAL 107
MY Y+F+++ E P + W ++T+ Q+ Q F+ + V F S C P + L
Sbjct 131 MYFYYFLSS--EYPGVRANIWWKKYITLTQLCQFFMLLSYAIYVRFFSP--NCGVP-RGL 185
Query 108 WHRQQLDDALSYGHYITLKNLYWACLMYSTYFYLFAEYFVKRYLRKPQ 155
LY + + YLF ++++ YLR P+
Sbjct 186 --------------------LYLNMVQGVVFIYLFGKFYIDNYLRPPK 213
> Hs8923312
Length=296
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 44/171 (25%), Positives = 73/171 (42%), Gaps = 38/171 (22%)
Query 2 LELIDTAFIILRKK--RLSFLHYYHHATVLLYTWDAYTCEQPAGIYFVA--MNYTVHAAM 57
+E +DT F +LRKK +++FLH YHHA+ + W P G F +N VH M
Sbjct 125 VEFLDTIFFVLRKKTSQITFLHVYHHAS-MFNIWWCVLNWIPCGQSFFGPTLNSFVHILM 183
Query 58 YLYFFIAA--VLERPLSWGIFVTVAQISQMFIGVGVTFVSMYIACTAPYKALWHRQQLDD 115
Y Y+ ++ + + L W ++T AQ+ Q + + T ++ C P+
Sbjct 184 YSYYGLSVFPSMHKYLWWKKYLTQAQLVQFVLTITHTMSAVVKPCGFPF----------- 232
Query 116 ALSYGHYITLKNLYWACLMYS-----TYFYLFAEYFVKRYLRKPQNNKHQQ 161
CL++ T LF ++V+ Y +KP Q+
Sbjct 233 ---------------GCLIFQSSYMLTLVILFLNFYVQTYRKKPMKKDMQE 268
> Hs13489093
Length=279
Score = 48.5 bits (114), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 34/91 (37%), Positives = 53/91 (58%), Gaps = 8/91 (8%)
Query 1 FLELIDTAFIILRKK--RLSFLHYYHHATVLLYTWDAYTCEQPAGI--YFVAMNYTVHAA 56
F+EL+DT ILRKK +++FLH +HH +VL ++W P G+ + +N +VH
Sbjct 119 FIELMDTVIFILRKKDGQVTFLHVFHH-SVLPWSWWWGVKIAPGGMGSFHAMINSSVHVI 177
Query 57 MYLYFFIAA---VLERPLSWGIFVTVAQISQ 84
MYLY+ ++A V + L W +T Q+ Q
Sbjct 178 MYLYYGLSAFGPVAQPYLWWKKHMTAIQLIQ 208
> HsM7705799
Length=200
Score = 48.1 bits (113), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 28/69 (40%), Positives = 44/69 (63%), Gaps = 5/69 (7%)
Query 1 FLELIDTAFIILRKK--RLSFLHYYHHATVLLYTWDAYTCEQPAGI--YFVAMNYTVHAA 56
F+EL+DT ILRKK +++FLH +HH +VL ++W P G+ + +N +VH
Sbjct 119 FIELMDTVIFILRKKDGQVTFLHVFHH-SVLPWSWWWGVKIAPGGMGSFHAMINSSVHVI 177
Query 57 MYLYFFIAA 65
MYLY+ ++A
Sbjct 178 MYLYYGLSA 186
> 7299264
Length=265
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 40/172 (23%), Positives = 79/172 (45%), Gaps = 31/172 (18%)
Query 1 FLELIDTAFIILRKK--RLSFLHYYHHATVLLYTWDAYTCEQPAGIYF--VAMNYTVHAA 56
F++L++T F +LRKK ++SFLH +HH + + T G F +N VH
Sbjct 116 FMDLLETVFFVLRKKDRQISFLHVFHHLVMSFGGYLHITFNGYGGTLFPLCLLNVAVHVI 175
Query 57 MYLYFFIAAVLE--RPLSWGIFVTVAQISQ-MFIGVGVTFVSMYIACTAPYKALWHRQQL 113
MY Y+++++V + + W ++T+ Q+ Q + + ++ M C A ++
Sbjct 176 MYAYYYLSSVSKDVQTSRWKKYITIVQLVQFILVLANFSYTLMQPDCNASRTVIYT---- 231
Query 114 DDALSYGHYITLKNLYWACLMYSTYFYLFAEYFVKRYLRKPQNNKHQQHQLQ 165
G +I+ +T+ +FA +++ Y+ N Q+ L+
Sbjct 232 ------GMFIS-----------TTFILMFANFYIHNYI---LNGSKQKSALK 263
> 7300878
Length=295
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 29/88 (32%), Positives = 49/88 (55%), Gaps = 6/88 (6%)
Query 3 ELIDTAFIILRK--KRLSFLHYYHHATVLLYTWDAYTCEQPA-GIYFVAMNYTVHAAMYL 59
EL+DT F +LRK ++++FLH YHH + + +W G + +N VH MY
Sbjct 124 ELLDTIFFVLRKNDRQVTFLHVYHHTVMPMISWGTSKYYPGGHGTFIGWINSFVHIIMYS 183
Query 60 YFFIAAV---LERPLSWGIFVTVAQISQ 84
Y+F++A +++ L W ++T Q+ Q
Sbjct 184 YYFLSAFGPQMQKYLWWKKYITNLQMIQ 211
> 7302564
Length=262
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 35/124 (28%), Positives = 61/124 (49%), Gaps = 7/124 (5%)
Query 2 LELIDTAFIILRK--KRLSFLHYYHHATVLLYTWDAYTCEQPAGIYFVA--MNYTVHAAM 57
++LIDT F +LRK K+++ LH YHH ++L Y P G Y + +N VH M
Sbjct 118 IDLIDTIFFVLRKSNKQITVLHVYHHVFMVLGVPLTYYFYGPGGQYNLMGYLNSFVHVVM 177
Query 58 YLYFFIAA---VLERPLSWGIFVTVAQISQMFIGVGVTFVSMYIACTAPYKALWHRQQLD 114
Y Y+F +A ++ W ++T Q Q I + +++++ + + QL
Sbjct 178 YAYYFASAWYPNVKSTFWWKEYITKLQFLQFMILFAQSVLTLWLNPGCRFPKVLQYVQLG 237
Query 115 DALS 118
++S
Sbjct 238 GSVS 241
> 7300874
Length=277
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 30/91 (32%), Positives = 49/91 (53%), Gaps = 6/91 (6%)
Query 3 ELIDTAFIILRKKR--LSFLHYYHHATVLLYTWDAYTCEQPA-GIYFVAMNYTVHAAMYL 59
EL+DT F +LRKK+ +SFLH YHH + + + G +N +H MY
Sbjct 129 ELLDTVFFVLRKKQRQISFLHLYHHTLMPVCAFIGVKYFAGGHGTLLGFINSFIHIIMYA 188
Query 60 YFFIAAV---LERPLSWGIFVTVAQISQMFI 87
Y+ ++A+ +++ L W ++T+ QI Q I
Sbjct 189 YYLLSAMGPKVQKYLWWKKYITILQIVQFLI 219
> 7296847
Length=245
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 32/91 (35%), Positives = 44/91 (48%), Gaps = 8/91 (8%)
Query 1 FLELIDTAFIILRKKR--LSFLHYYHHATVLLYTWDAYTCEQPAG--IYFVAMNYTVHAA 56
F E DT F ILRKK +S LH HH + W P G +F +N VH
Sbjct 15 FTEFFDTLFFILRKKNEHVSTLHVIHHGCMPFSVWMGLKFA-PGGHSTFFALLNSFVHIV 73
Query 57 MYLYFFIAAV---LERPLSWGIFVTVAQISQ 84
MY Y+ IAA+ ++ + W ++T Q+ Q
Sbjct 74 MYFYYMIAAMGPKYQKYIWWKKYLTTFQMVQ 104
> 7294766
Length=140
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 24/44 (54%), Positives = 29/44 (65%), Gaps = 3/44 (6%)
Query 2 LELIDTAFIILRKK--RLSFLHYYHHATVLLYTWDAYTCEQPAG 43
LE +DTAF ILR K +LSFLH YHH+T+ L+ W Y P G
Sbjct 16 LEFVDTAFFILRHKWNQLSFLHVYHHSTMFLFCW-TYVKWLPTG 58
> 7300933
Length=272
Score = 42.4 bits (98), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 32/94 (34%), Positives = 50/94 (53%), Gaps = 15/94 (15%)
Query 1 FLELIDTAFIILRKK--RLSFLHYYHHA------TVLLYTWDAYTCEQPAGIYFVAMNYT 52
+L+L+DT FI+LRKK ++SFLH YHH ++ + C GI +N
Sbjct 123 YLDLLDTVFIVLRKKNSQVSFLHVYHHGGMVFGVSIFMTFLGGSHCSM-LGI----INLL 177
Query 53 VHAAMYLYFFIAAV--LERPLSWGIFVTVAQISQ 84
VH MY Y++ A++ ++ L W +T Q+ Q
Sbjct 178 VHTVMYAYYYAASLGAVKNLLWWKQRITQLQLMQ 211
> 7299268
Length=264
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 30/96 (31%), Positives = 52/96 (54%), Gaps = 7/96 (7%)
Query 2 LELIDTAFIILRKK--RLSFLHYYHHATVLLYTWDAYTCEQPAGIYFVA--MNYTVHAAM 57
L+L++T F +LRKK ++SFLH +HH +L +++ G F N VH M
Sbjct 118 LDLLETVFFVLRKKDRQISFLHVFHHMYMLYFSFMYLYYYGYGGHGFFMCFFNVVVHIMM 177
Query 58 YLYFFIAAV---LERPLSWGIFVTVAQISQMFIGVG 90
Y Y++ +++ + L W ++T+ Q+ Q I +G
Sbjct 178 YSYYYQSSLNRDSKGDLWWKKYITIVQLIQFGIVLG 213
> 7299266
Length=262
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 42/166 (25%), Positives = 80/166 (48%), Gaps = 28/166 (16%)
Query 2 LELIDTAFIILRKK--RLSFLHYYHHATVLLYTWDAYTCEQPAG-IYFVAM-NYTVHAAM 57
L+L+DT F +LRKK +++FLH +HH ++ + G ++ + M N VH M
Sbjct 114 LDLMDTIFFVLRKKQRQITFLHVFHHVFMVFTSHMLIRFYGFGGHVFLICMFNVLVHIVM 173
Query 58 YLYFFIAA---VLERPLSWGIFVTVAQISQMFIGVGVTFVSMYIACTAPYKALWHRQQLD 114
Y Y++ ++ ++ L W ++T+ Q+ Q F+ M++ C Y Q +
Sbjct 174 YGYYYASSQSQNVQESLWWKKYLTLGQLVQ--------FLLMFLHCMYTYF------QPN 219
Query 115 DALSYGHYITLKNLYWACLMYSTYFYLFAEYFVKRYLRKPQNNKHQ 160
+ S G +Y + F +F ++++K Y+R P+ K +
Sbjct 220 CSASRG------VIYVISSASAFMFLMFTKFYIKTYIR-PKEVKSK 258
> 7300879
Length=262
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 30/86 (34%), Positives = 43/86 (50%), Gaps = 6/86 (6%)
Query 3 ELIDTAFIILRKK--RLSFLHYYHHATVLLYTWDAYTCEQPAGIYFVA-MNYTVHAAMYL 59
E DT F +LRKK ++++LH YHH+ L TW F +N VH MY
Sbjct 157 EFADTMFFVLRKKSSQITWLHVYHHSVTPLETWVLVKFLAGGNATFPNLLNNFVHVCMYF 216
Query 60 YFFIAAV---LERPLSWGIFVTVAQI 82
Y+ +AA+ + L W ++T QI
Sbjct 217 YYMMAAMGPEYAKFLWWKKYMTELQI 242
> 7299267
Length=261
Score = 37.4 bits (85), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 28/93 (30%), Positives = 49/93 (52%), Gaps = 7/93 (7%)
Query 2 LELIDTAFIILRKK--RLSFLHYYHHATV--LLYTWDAYTCEQPAGIYFVAMNYTVHAAM 57
++L++T F I RKK ++SFLH +HH + + + + Y G + + N VH M
Sbjct 114 MDLLETVFFIFRKKYRQISFLHVFHHVYMVYIGFLYMYYYGYGGHGFFLITFNVVVHTMM 173
Query 58 YLYFFIAAVLERP---LSWGIFVTVAQISQMFI 87
Y Y++ +++ L W ++TV Q+ Q I
Sbjct 174 YTYYYQSSLNRNSGGDLWWKKYITVVQLVQFVI 206
> 7298969
Length=276
Score = 33.1 bits (74), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 34/167 (20%), Positives = 74/167 (44%), Gaps = 28/167 (16%)
Query 11 ILRKK--RLSFLHYYHHATVLLYTWDAYTCEQPAG--IYFVAMNYTVHAAMYLYFFIAAV 66
++RK+ ++S LH HH + + W P G +F +N VH MY Y+ +AA+
Sbjct 80 VMRKRYDQVSTLHVIHHGIMPVSVWWGVKFT-PGGHSTFFGFLNTFVHIFMYAYYMLAAM 138
Query 67 ---LERPLSWGIFVTVAQISQMFIGVGVTFVSMYIACTAPYKALWHRQQLDDALSYGHYI 123
+++ L W ++TV Q+ Q + + +F L+ + + + + ++I
Sbjct 139 GPKVQKYLWWKKYLTVMQMIQFVLVMVHSF------------QLFFKNDCNYPIGFAYFI 186
Query 124 TLKNLYWACLMYSTYFYLFAEYFVKRYLRKPQNNKHQQHQLQHPKEH 170
+ +++LF+ ++ + Y+++ +K H H
Sbjct 187 GAHAV--------MFYFLFSNFYKRAYVKRDGKDKASVKANGHANGH 225
> At1g72120
Length=1095
Score = 28.5 bits (62), Expect = 7.7, Method: Composition-based stats.
Identities = 19/67 (28%), Positives = 33/67 (49%), Gaps = 4/67 (5%)
Query 44 IYFVAMNYTVHAAMYLYFFI--AAVLERPLSWGIFVTVAQISQMFIGVG-VTFVSMYIAC 100
++F + Y + A Y+ FF ++R + G+ + A + Q+FIG+ V FV +Y
Sbjct 322 VWFTTLAYAIPYAQYMTFFTKQGVTMDRTILPGVKIPPASL-QVFIGISIVLFVPIYDRV 380
Query 101 TAPYKAL 107
P L
Sbjct 381 FVPIARL 387
Lambda K H
0.331 0.140 0.461
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2598880752
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40