bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0811_orf1
Length=111
Score E
Sequences producing significant alignments: (Bits) Value
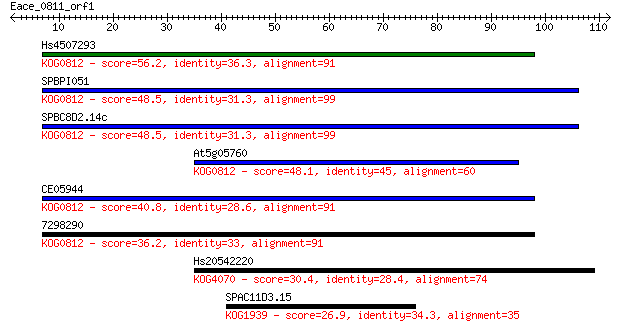
Hs4507293 56.2 2e-08
SPBPI051 48.5 3e-06
SPBC8D2.14c 48.5 3e-06
At5g05760 48.1 4e-06
CE05944 40.8 6e-04
7298290 36.2 0.015
Hs20542220 30.4 0.76
SPAC11D3.15 26.9 9.9
> Hs4507293
Length=301
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 33/92 (35%), Positives = 54/92 (58%), Gaps = 1/92 (1%)
Query 7 DRTEEFLAAAESFQLPGGGAQNDTQRHFSDADR-HFNALASEMGNALHATSLKLQELGKL 65
DRT+EFL+A +S Q G Q + + R F +A +G L T KL++L L
Sbjct 5 DRTQEFLSACKSLQTRQNGIQTNKPALRAVRQRSEFTLMAKRIGKDLSNTFAKLEKLTIL 64
Query 66 ARQCGIYNVKTVQIPELTFDIRKTINALNCKV 97
A++ +++ K V+I ELT+ I++ IN+LN ++
Sbjct 65 AKRKSLFDDKAVEIEELTYIIKQDINSLNKQI 96
> SPBPI051
Length=309
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 31/109 (28%), Positives = 56/109 (51%), Gaps = 16/109 (14%)
Query 7 DRTEEF----------LAAAESFQLPGGGAQNDTQRHFSDADRHFNALASEMGNALHATS 56
DRT EF L + Q GG Q Q+ F +A ++ N ++ T
Sbjct 5 DRTAEFQACVTKTRSRLRTTTANQAVGGPDQTKHQKS------EFTRIAQKIANQINQTG 58
Query 57 LKLQELGKLARQCGIYNVKTVQIPELTFDIRKTINALNCKVDVFARAVE 105
KLQ+L +LA++ +++ + V+I ELTF I++++++LN + + V+
Sbjct 59 EKLQKLSQLAKRKTLFDDRPVEIQELTFQIKQSLSSLNSDIASLQQVVK 107
> SPBC8D2.14c
Length=309
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 31/109 (28%), Positives = 56/109 (51%), Gaps = 16/109 (14%)
Query 7 DRTEEF----------LAAAESFQLPGGGAQNDTQRHFSDADRHFNALASEMGNALHATS 56
DRT EF L + Q GG Q Q+ F +A ++ N ++ T
Sbjct 5 DRTAEFQACVTKTRSRLRTTTANQAVGGPDQTKHQKS------EFTRIAQKIANQINQTG 58
Query 57 LKLQELGKLARQCGIYNVKTVQIPELTFDIRKTINALNCKVDVFARAVE 105
KLQ+L +LA++ +++ + V+I ELTF I++++++LN + + V+
Sbjct 59 EKLQKLSQLAKRKTLFDDRPVEIQELTFQIKQSLSSLNSDIASLQQVVK 107
> At5g05760
Length=336
Score = 48.1 bits (113), Expect = 4e-06, Method: Composition-based stats.
Identities = 27/60 (45%), Positives = 34/60 (56%), Gaps = 0/60 (0%)
Query 35 SDADRHFNALASEMGNALHATSLKLQELGKLARQCGIYNVKTVQIPELTFDIRKTINALN 94
S FN AS +G + TS K+ L KLA+Q I+N +TV+I ELT IR I LN
Sbjct 41 SSPGSEFNKKASRIGLGIKETSQKITRLAKLAKQSTIFNDRTVEIQELTVLIRNDITGLN 100
> CE05944
Length=413
Score = 40.8 bits (94), Expect = 6e-04, Method: Composition-based stats.
Identities = 26/93 (27%), Positives = 46/93 (49%), Gaps = 3/93 (3%)
Query 7 DRTEEFLAAAESFQLPGG--GAQNDTQRHFSDADRHFNALASEMGNALHATSLKLQELGK 64
DRT EF A A+S+++ G + + FN LA +G L T K+++L +
Sbjct 111 DRTSEFRATAKSYEMKAAANGIRPQPKHEMLSESVQFNQLAKRIGKELSQTCAKMEKLAE 170
Query 65 LARQCGIYNVKTVQIPELTFDIRKTINALNCKV 97
A++ Y ++ QI L+ ++ I LN ++
Sbjct 171 YAKKKSCYEERS-QIDHLSSIVKSDITGLNKQI 202
> 7298290
Length=310
Score = 36.2 bits (82), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 30/100 (30%), Positives = 52/100 (52%), Gaps = 14/100 (14%)
Query 7 DRTEEFLAAAESFQLPGGGAQNDTQR-HFSDADR--------HFNALASEMGNALHATSL 57
DRT EF A S Q A+N T+ + D + F +A +G + +T
Sbjct 5 DRTGEFANAIRSLQ-----ARNITRAVNIRDPRKAKQVQSYSEFMMVARFIGKNIASTYA 59
Query 58 KLQELGKLARQCGIYNVKTVQIPELTFDIRKTINALNCKV 97
KL++L LA++ +++ + +I ELT+ I+ +NALN ++
Sbjct 60 KLEKLTMLAKKKSLFDDRPQEIQELTYIIKGDLNALNQQI 99
> Hs20542220
Length=170
Score = 30.4 bits (67), Expect = 0.76, Method: Compositional matrix adjust.
Identities = 21/76 (27%), Positives = 35/76 (46%), Gaps = 2/76 (2%)
Query 35 SDADRHFNALASEMGNALHATSLKLQELGKLARQCGIYNVKTVQIP--ELTFDIRKTINA 92
S+A++ F+ A+ ++ T + + KL + CGI + KTV ++ F K NA
Sbjct 3 SEAEKTFHRFAAFGESSSSGTEMNNKNFSKLCKDCGIMDGKTVTSTDVDIVFSKVKAKNA 62
Query 93 LNCKVDVFARAVENAG 108
F AV+ G
Sbjct 63 RTITFQQFKEAVKELG 78
> SPAC11D3.15
Length=1317
Score = 26.9 bits (58), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 12/35 (34%), Positives = 19/35 (54%), Gaps = 0/35 (0%)
Query 41 FNALASEMGNALHATSLKLQELGKLARQCGIYNVK 75
F A+A +MG AL TS+ +L C +++ K
Sbjct 760 FMAVAEQMGRALQKTSVSTNVKERLDYSCALFDAK 794
Lambda K H
0.317 0.133 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1192473466
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40