bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0819_orf1
Length=160
Score E
Sequences producing significant alignments: (Bits) Value
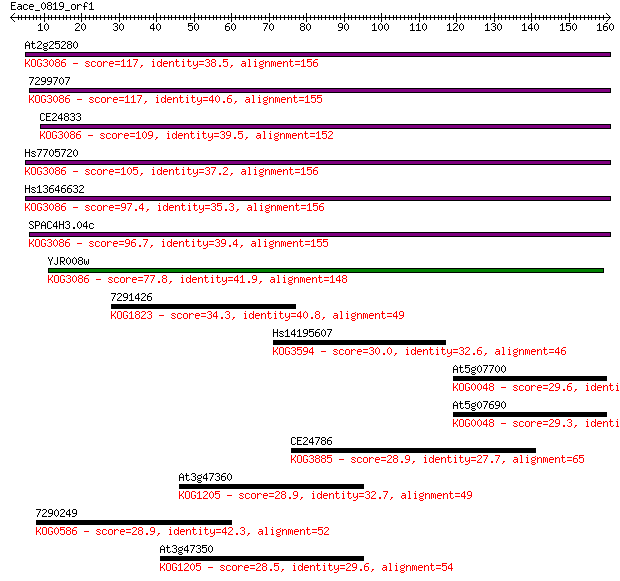
At2g25280 117 8e-27
7299707 117 1e-26
CE24833 109 2e-24
Hs7705720 105 3e-23
Hs13646632 97.4 1e-20
SPAC4H3.04c 96.7 2e-20
YJR008w 77.8 7e-15
7291426 34.3 0.099
Hs14195607 30.0 1.8
At5g07700 29.6 2.9
At5g07690 29.3 3.0
CE24786 28.9 4.1
At3g47360 28.9 4.4
7290249 28.9 4.5
At3g47350 28.5 6.0
> At2g25280
Length=291
Score = 117 bits (294), Expect = 8e-27, Method: Compositional matrix adjust.
Identities = 60/157 (38%), Positives = 91/157 (57%), Gaps = 4/157 (2%)
Query 5 SAGVYLTPLGDLPLDTDTLQALRLTGAFCEASISEDEEEHSIELMLPFVRHTF-SEGVKI 63
+A VY TP+G+LP+D + ++ +R G F + DE EHS+E+ LP++ F VK+
Sbjct 90 TATVYKTPIGNLPVDVEMIKEIRAMGKFGMMDLRVDEAEHSMEMHLPYLAKVFEGNNVKV 149
Query 64 VPMIVGDLKSLEDCQKYADLLINYFLQDENLFIVSSDFCHWGPRYSYFYLEDPQPGVPIH 123
VP++VG + S E+ Y +LL Y +N F VSSDFCHWG R++Y + ++ IH
Sbjct 150 VPILVGAV-SPENEAMYGELLAKYVDDPKNFFSVSSDFCHWGSRFNYMHYDNTHGA--IH 206
Query 124 AAIEKMDRKAVKFITEHQGQGFYDYLESTGLSVCGRH 160
+IE +D+K + I F YL ++CGRH
Sbjct 207 KSIEALDKKGMDIIETGDPDAFKKYLLEFENTICGRH 243
> 7299707
Length=295
Score = 117 bits (292), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 63/158 (39%), Positives = 88/158 (55%), Gaps = 6/158 (3%)
Query 6 AGVYLTPLGDLPLDTDTLQALRLTGAFCEASISEDEEEHSIELMLPFVRHT---FSEGVK 62
A Y TPL DL +D L TG F + DE+EHSIE+ LP++ + +
Sbjct 89 AKKYRTPLYDLKIDAQINSELEKTGKFSWMDMKTDEDEHSIEMHLPYIAKVMEDYKDQFT 148
Query 63 IVPMIVGDLKSLEDCQKYADLLINYFLQDENLFIVSSDFCHWGPRYSYFYLEDPQPGVPI 122
IVP++VG L ++ Q Y LL +Y + NLF++SSDFCHWG R+SY Y D G I
Sbjct 149 IVPILVGSLNPEQEAQ-YGSLLSSYLMDPTNLFVISSDFCHWGHRFSYTYY-DSSCGA-I 205
Query 123 HAAIEKMDRKAVKFITEHQGQGFYDYLESTGLSVCGRH 160
H +IEK+D++ + I F +YL ++CGRH
Sbjct 206 HKSIEKLDKQGMDIIESLNPHSFTEYLRKYNNTICGRH 243
> CE24833
Length=392
Score = 109 bits (273), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 60/153 (39%), Positives = 84/153 (54%), Gaps = 3/153 (1%)
Query 9 YLTPLGDLPLDTDTLQALRLTGAFCEASISEDEEEHSIELMLPFVRHTF-SEGVKIVPMI 67
Y TPLGDL +D + LR T F ++E EHSIE+ LPF+ S+ IVP++
Sbjct 189 YRTPLGDLIVDHKINEELRATRHFDLMDRRDEESEHSIEMQLPFIAKVMGSKRYTIVPVL 248
Query 68 VGDLKSLEDCQKYADLLINYFLQDENLFIVSSDFCHWGPRYSYFYLEDPQPGVPIHAAIE 127
VG L Q Y ++ +Y NLF++SSDFCHWG R+S F D +PI+ I
Sbjct 249 VGSLPGSRQ-QTYGNIFAHYMEDPRNLFVISSDFCHWGERFS-FSPYDRHSSIPIYEQIT 306
Query 128 KMDRKAVKFITEHQGQGFYDYLESTGLSVCGRH 160
MD++ + I F DYL+ T ++CGR+
Sbjct 307 NMDKQGMSAIETLNPAAFNDYLKKTQNTICGRN 339
> Hs7705720
Length=297
Score = 105 bits (263), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 58/159 (36%), Positives = 86/159 (54%), Gaps = 6/159 (3%)
Query 5 SAGVYLTPLGDLPLDTDTLQALRLTGAFCEASISEDEEEHSIELMLPFVRHTF---SEGV 61
S +Y TPL DL +D L TG F S+ DE+EHSIE+ LP+ +
Sbjct 92 SVDIYRTPLYDLRIDQKIYGELWKTGMFERMSLQTDEDEHSIEMHLPYTAKAMESHKDEF 151
Query 62 KIVPMIVGDLKSLEDCQKYADLLINYFLQDENLFIVSSDFCHWGPRYSYFYLEDPQPGVP 121
I+P++VG L ++ Q++ L Y NLF+VSSDFCHWG R+ Y Y ++ Q
Sbjct 152 TIIPVLVGALSESKE-QEFGKLFSKYLADPSNLFVVSSDFCHWGQRFRYSYYDESQG--E 208
Query 122 IHAAIEKMDRKAVKFITEHQGQGFYDYLESTGLSVCGRH 160
I+ +IE +D+ + I + F +YL+ ++CGRH
Sbjct 209 IYRSIEHLDKMGMSIIEQLDPVSFSNYLKKYHNTICGRH 247
> Hs13646632
Length=295
Score = 97.4 bits (241), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 55/159 (34%), Positives = 83/159 (52%), Gaps = 8/159 (5%)
Query 5 SAGVYLTPLGDLPLDTDTLQALRLTGAFCEASISEDEEEHSIELMLPFVRHTF---SEGV 61
S +Y TPL DL +D L TG F S+ DE+E SIE+ LP+ +
Sbjct 92 SVDIYRTPLYDLRIDQKIYGELWKTGMFERMSLQTDEDERSIEMHLPYTAKAMESHKDEF 151
Query 62 KIVPMIVGDLKSLEDCQKYADLLINYFLQDENLFIVSSDFCHWGPRYSYFYLEDPQPGVP 121
I+P++VG +L + ++L Y NLF+VSSDFCHWG R+ + Y ++ Q
Sbjct 152 TIIPLLVG---ALSQKNRNSELFSKYLADPSNLFVVSSDFCHWGQRFRHSYYDESQG--E 206
Query 122 IHAAIEKMDRKAVKFITEHQGQGFYDYLESTGLSVCGRH 160
I +IE +D+ + I + F +YL+ ++CGRH
Sbjct 207 ICRSIEHLDKMGMSIIEQLDPVSFSNYLKKYHNTICGRH 245
> SPAC4H3.04c
Length=309
Score = 96.7 bits (239), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 61/177 (34%), Positives = 88/177 (49%), Gaps = 23/177 (12%)
Query 6 AGVYLTPLGDLPLDTDTLQALRLT-GAFCEASISEDEEEHSIELMLPFVR-HTFSEG--- 60
A + TPLGDL +D D Q L + +F ++ DE EHS+E+ P + H +G
Sbjct 90 ASICSTPLGDLKVDEDLCQKLVASDNSFDSMTLDVDESEHSLEMQFPLLAFHLLKQGCLG 149
Query 61 -VKIVPMIVGDLKSLEDCQKYADLLINYFLQDENLFIVSSDFCHWGPRYSY-FYLEDPQ- 117
VKIVP+++G L S A L Y + N F++SSDFCHWG R+ Y YL D
Sbjct 150 KVKIVPIMIGALTS-TTMMAAAKFLSQYIKDESNSFVISSDFCHWGRRFGYTLYLNDTNQ 208
Query 118 -------------PGVP-IHAAIEKMDRKAVKFITEHQGQGFYDYLESTGLSVCGRH 160
P P I+ +I +D +K I F +YL++T ++CGR+
Sbjct 209 LEDAVLKYKRRGGPTSPKIYESISNLDHIGMKIIETKSSDDFSEYLKTTQNTICGRY 265
> YJR008w
Length=338
Score = 77.8 bits (190), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 62/189 (32%), Positives = 84/189 (44%), Gaps = 42/189 (22%)
Query 11 TPLGDLPLDTDTLQAL-------RLTGAFCEASISEDEEEHSIELMLPFVRHTFS----- 58
TPLG+L +DTD + L F D EHS+E+ LP + T
Sbjct 98 TPLGNLKVDTDLCKTLIQKEYPENGKKLFKPMDHDTDMAEHSLEMQLPMLVETLKWREIS 157
Query 59 -EGVKIVPMIVGDLKSLEDCQKYADLLINYFLQDENLFIVSSDFCHWGPRYSYF------ 111
+ VK+ PM+V S++ + ++L Y NLFIVSSDFCHWG R+ Y
Sbjct 158 LDTVKVFPMMVSH-NSVDVDRCIGNILSEYIKDPNNLFIVSSDFCHWGRRFQYTGYVGSK 216
Query 112 -YLEDP-----------------QPGVPIHAAIEKMDRKAVKFITEHQGQGFYD----YL 149
L D VPI +IE MDR A+K +++ YD YL
Sbjct 217 EELNDAIQEETEVEMLTARSKLSHHQVPIWQSIEIMDRYAMKTLSDTPNGERYDAWKQYL 276
Query 150 ESTGLSVCG 158
E TG ++CG
Sbjct 277 EITGNTICG 285
> 7291426
Length=389
Score = 34.3 bits (77), Expect = 0.099, Method: Compositional matrix adjust.
Identities = 20/50 (40%), Positives = 27/50 (54%), Gaps = 1/50 (2%)
Query 28 LTGAFCEASISED-EEEHSIELMLPFVRHTFSEGVKIVPMIVGDLKSLED 76
L C A+ SE +E+ I+LMLP V H V +P++VG L L D
Sbjct 298 LWQKLCSAASSESVDEQRVIDLMLPLVTHKDGRYVSELPLVVGTLLKLLD 347
> Hs14195607
Length=818
Score = 30.0 bits (66), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 15/46 (32%), Positives = 22/46 (47%), Gaps = 0/46 (0%)
Query 71 LKSLEDCQKYADLLINYFLQDENLFIVSSDFCHWGPRYSYFYLEDP 116
L+S +D + L NY L F + + FC GP+Y+ L P
Sbjct 154 LQSAQDLDVGLNGLQNYTLSANGYFHLHTRFCSHGPKYAELVLNKP 199
> At5g07700
Length=338
Score = 29.6 bits (65), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 13/41 (31%), Positives = 21/41 (51%), Gaps = 0/41 (0%)
Query 119 GVPIHAAIEKMDRKAVKFITEHQGQGFYDYLESTGLSVCGR 159
G+ A + D+K + +I +H G+ D E GL CG+
Sbjct 11 GLKKGAWTTEEDKKLISYIHDHGEGGWRDIPEKAGLKRCGK 51
> At5g07690
Length=336
Score = 29.3 bits (64), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 13/41 (31%), Positives = 21/41 (51%), Gaps = 0/41 (0%)
Query 119 GVPIHAAIEKMDRKAVKFITEHQGQGFYDYLESTGLSVCGR 159
G+ A + D+K + +I EH G+ D + GL CG+
Sbjct 11 GLKKGAWTAEEDKKLISYIHEHGEGGWRDIPQKAGLKRCGK 51
> CE24786
Length=425
Score = 28.9 bits (63), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 18/65 (27%), Positives = 31/65 (47%), Gaps = 8/65 (12%)
Query 76 DCQKYADLLINYFLQDENLFIVSSDFCHWGPRYSYFYLEDPQPGVPIHAAIEKMDRKAVK 135
+C +++ NY+ NL+ C +G R++ +Y+E + G P K RKA
Sbjct 154 ECVFLEEMMENYY----NLYAS----CAYGDRFNPWYIELRRSGKPRRGPNSKKRRKASH 205
Query 136 FITEH 140
F+ H
Sbjct 206 FLVVH 210
> At3g47360
Length=309
Score = 28.9 bits (63), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 29/49 (59%), Gaps = 3/49 (6%)
Query 46 IELMLPFVRHTFSEGVKIVPMIVGDLKSLEDCQKYADLLINYFLQDENL 94
+E++ R S V I+P GD+ ++EDC+K+ D I +F + ++L
Sbjct 84 LEIVAETSRQLGSGNVIIIP---GDVSNVEDCKKFIDETIRHFGKLDHL 129
> 7290249
Length=1398
Score = 28.9 bits (63), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 22/58 (37%), Positives = 28/58 (48%), Gaps = 6/58 (10%)
Query 8 VYLTPLGDLPLDTDTLQALR---LTGAFCEASISEDEEEHSIELML---PFVRHTFSE 59
+Y+ G LP D TLQ+LR L+G F E EH I ML P R+T +
Sbjct 328 LYVLVCGALPFDGSTLQSLRDRVLSGRFRIPFFMSSECEHLIRRMLVLEPTRRYTIDQ 385
> At3g47350
Length=308
Score = 28.5 bits (62), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 16/54 (29%), Positives = 31/54 (57%), Gaps = 3/54 (5%)
Query 41 EEEHSIELMLPFVRHTFSEGVKIVPMIVGDLKSLEDCQKYADLLINYFLQDENL 94
+ +E++ R S V I+P GD+ ++EDC+K+ D I++F + ++L
Sbjct 78 RRKDRLEIVAETSRQLGSGDVIIIP---GDVSNVEDCKKFIDETIHHFGKLDHL 128
Lambda K H
0.321 0.140 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2172509160
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40