bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
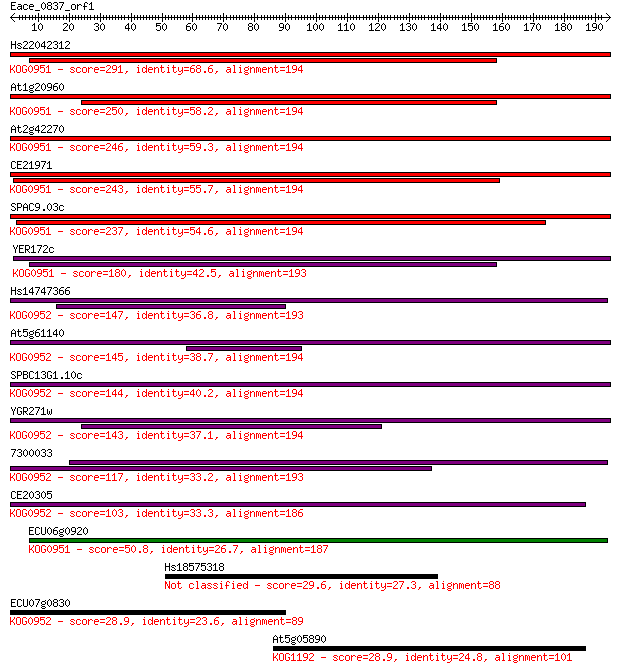
Query= Eace_0837_orf1
Length=194
Score E
Sequences producing significant alignments: (Bits) Value
Hs22042312 291 6e-79
At1g20960 250 1e-66
At2g42270 246 2e-65
CE21971 243 1e-64
SPAC9.03c 237 1e-62
YER172c 180 1e-45
Hs14747366 147 1e-35
At5g61140 145 6e-35
SPBC13G1.10c 144 9e-35
YGR271w 143 3e-34
7300033 117 2e-26
CE20305 103 2e-22
ECU06g0920 50.8 2e-06
Hs18575318 29.6 3.9
ECU07g0830 28.9 6.1
At5g05890 28.9 6.1
> Hs22042312
Length=2136
Score = 291 bits (745), Expect = 6e-79, Method: Compositional matrix adjust.
Identities = 133/194 (68%), Positives = 163/194 (84%), Gaps = 0/194 (0%)
Query 1 EEEKLELQRLMERVPIPVKGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANR 60
EEEKLELQ+L+ERVPIPVK S +EPS+K+NVLLQA+IS+LKLEGFA+MADMVYV QSA R
Sbjct 1031 EEEKLELQKLLERVPIPVKESIEEPSAKINVLLQAFISQLKLEGFALMADMVYVTQSAGR 1090
Query 61 IMRAIFDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLPF 120
+MRAIF+I L RGWAQL + L LC I RMW +M PLRQF+ LPEE+++KIEKK+ PF
Sbjct 1091 LMRAIFEIVLNRGWAQLTDKTLNLCKMIDKRMWQSMCPLRQFRKLPEEVVKKIEKKNFPF 1150
Query 121 DRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELAAFVQPLSRSCLVVELTITPDFQW 180
+R YDL EIGEL+R+PKMGK +H+ +H FPKLEL+ +QP++RS L VELTITPDFQW
Sbjct 1151 ERLYDLNHNEIGELIRMPKMGKTIHKYVHLFPKLELSVHLQPITRSTLKVELTITPDFQW 1210
Query 181 DQKIHGNGEVFWIL 194
D+K+HG+ E FWIL
Sbjct 1211 DEKVHGSSEAFWIL 1224
Score = 52.4 bits (124), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 38/164 (23%), Positives = 78/164 (47%), Gaps = 24/164 (14%)
Query 7 LQRLMERVPIPVKGSP-DEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANRIMRAI 65
L++L ++VP + ++P K N+LLQA++S+++L + +D + A R+++A
Sbjct 1868 LRQLAQKVPHKLNNPKFNDPHVKTNLLLQAHLSRMQLSA-ELQSDTEEILSKAIRLIQAC 1926
Query 66 FDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRK------------I 113
D+ GW A+ A++L + MWS + L+Q E +++ +
Sbjct 1927 VDVLSSNGWLSPALAAMELAQMVTQAMWSKDSYLKQLPHFTSEHIKRCTDKGVESVFDIM 1986
Query 114 EKKDLPFDRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELA 157
E +D + LT ++I ++ R + +P +EL+
Sbjct 1987 EMEDEERNALLQLTDSQIADVA----------RFCNRYPNIELS 2020
> At1g20960
Length=2171
Score = 250 bits (639), Expect = 1e-66, Method: Compositional matrix adjust.
Identities = 113/194 (58%), Positives = 157/194 (80%), Gaps = 0/194 (0%)
Query 1 EEEKLELQRLMERVPIPVKGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANR 60
++EK+EL +L++RVPIP+K + +EPS+K+NVLLQAYIS+LKLEG ++ +DMVY+ QSA R
Sbjct 1055 QDEKMELAKLLDRVPIPIKETLEEPSAKINVLLQAYISQLKLEGLSLTSDMVYITQSAGR 1114
Query 61 IMRAIFDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLPF 120
++RA+++I L+RGWAQLA +AL L + RMWS TPLRQF L +++ ++EKKDL +
Sbjct 1115 LVRALYEIVLKRGWAQLAEKALNLSKMVGKRMWSVQTPLRQFHGLSNDILMQLEKKDLVW 1174
Query 121 DRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELAAFVQPLSRSCLVVELTITPDFQW 180
+RYYDL++ E+GEL+R PKMGK LH+ IH FPK+ L+A VQP++R+ L VELT+TPDF W
Sbjct 1175 ERYYDLSAQELGELIRSPKMGKPLHKFIHQFPKVTLSAHVQPITRTVLNVELTVTPDFLW 1234
Query 181 DQKIHGNGEVFWIL 194
D+KIH E FWI+
Sbjct 1235 DEKIHKYVEPFWII 1248
Score = 52.0 bits (123), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 38/139 (27%), Positives = 64/139 (46%), Gaps = 6/139 (4%)
Query 24 EPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANRIMRAIFDICLRRGWAQLAIRALQ 83
+P K N LLQA+ S+ + G M D V SA R+++A+ D+ GW LA+ A++
Sbjct 1913 DPHVKANALLQAHFSRQNIGGNLAM-DQRDVLLSATRLLQAMVDVISSNGWLNLALLAME 1971
Query 84 LCNEIQGRMWSTMTPLRQFKVLPEELMRKI-EKKDLPFDRYYDLTSTEIGELVRVPKMGK 142
+ + MW + L Q ++L ++ E + +DL E E + KM
Sbjct 1972 VSQMVTQGMWERDSMLLQLPHFTKDLAKRCQENPGKNIETVFDLVEMEDEERQELLKMSD 2031
Query 143 L----LHRLIHSFPKLELA 157
+ R + FP ++L
Sbjct 2032 AQLLDIARFCNRFPNIDLT 2050
> At2g42270
Length=2172
Score = 246 bits (628), Expect = 2e-65, Method: Compositional matrix adjust.
Identities = 115/194 (59%), Positives = 154/194 (79%), Gaps = 0/194 (0%)
Query 1 EEEKLELQRLMERVPIPVKGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANR 60
++EK+EL +L++RVPIPVK + ++PS+K+NVLLQ YISKLKLEG ++ +DMVY+ QSA R
Sbjct 1056 QDEKMELAKLLDRVPIPVKETLEDPSAKINVLLQVYISKLKLEGLSLTSDMVYITQSAGR 1115
Query 61 IMRAIFDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLPF 120
++RAIF+I L+RGWAQL+ +AL L + RMWS TPL QF +P+E++ K+EK DL +
Sbjct 1116 LLRAIFEIVLKRGWAQLSQKALNLSKMVGKRMWSVQTPLWQFPGIPKEILMKLEKNDLVW 1175
Query 121 DRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELAAFVQPLSRSCLVVELTITPDFQW 180
+RYYDL+S E+GEL+ PKMG+ LH+ IH FPKL+LAA VQP+SRS L VELT+TPDF W
Sbjct 1176 ERYYDLSSQELGELICNPKMGRPLHKYIHQFPKLKLAAHVQPISRSVLQVELTVTPDFHW 1235
Query 181 DQKIHGNGEVFWIL 194
D K + E FWI+
Sbjct 1236 DDKANKYVEPFWII 1249
> CE21971
Length=2145
Score = 243 bits (621), Expect = 1e-64, Method: Compositional matrix adjust.
Identities = 108/194 (55%), Positives = 147/194 (75%), Gaps = 0/194 (0%)
Query 1 EEEKLELQRLMERVPIPVKGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANR 60
+EEKLELQ++ E PIP+K + DE S+K NVLLQAYIS+LKLEGFA+ ADMV+V QSA R
Sbjct 1024 DEEKLELQKMAEHAPIPIKENLDEASAKTNVLLQAYISQLKLEGFALQADMVFVAQSAGR 1083
Query 61 IMRAIFDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLPF 120
+ RA+F+I L RGWA LA + L LC + R W ++ PL QFK +P E++R I+KK+ F
Sbjct 1084 LFRALFEIVLWRGWAGLAQKVLTLCKMVTQRQWGSLNPLHQFKKIPSEVVRSIDKKNYSF 1143
Query 121 DRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELAAFVQPLSRSCLVVELTITPDFQW 180
DR YDL ++G+L+++PKMGK L + I FPKLE+ +QP++R+ + +ELTITPDF+W
Sbjct 1144 DRLYDLDQHQLGDLIKMPKMGKPLFKFIRQFPKLEMTTLIQPITRTTMRIELTITPDFKW 1203
Query 181 DQKIHGNGEVFWIL 194
D+K+HG+ E FWI
Sbjct 1204 DEKVHGSAEGFWIF 1217
Score = 53.1 bits (126), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 44/167 (26%), Positives = 77/167 (46%), Gaps = 18/167 (10%)
Query 2 EEKLELQRLMERVPIPVKGSP-DEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANR 60
+E + L++L ER+P +K +P KVN+L+ A++S++KL + D + A R
Sbjct 1862 KEDVILRQLAERLPGQLKNQKFTDPHVKVNLLIHAHLSRVKLTA-ELNKDTELIVLRACR 1920
Query 61 IMRAIFDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKK---- 116
+++A D+ GW AI A++L + M+S L+Q L+ + + K
Sbjct 1921 LVQACVDVLSSNGWLSPAIHAMELSQMLTQAMYSNEPYLKQLPHCSAALLERAKAKEVTS 1980
Query 117 -----DLPFDRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELAA 158
+L D D+ E EL V R + +P +E+A
Sbjct 1981 VFELLELENDDRSDILQMEGAELADVA-------RFCNHYPSIEVAT 2020
> SPAC9.03c
Length=2176
Score = 237 bits (605), Expect = 1e-62, Method: Compositional matrix adjust.
Identities = 106/194 (54%), Positives = 146/194 (75%), Gaps = 0/194 (0%)
Query 1 EEEKLELQRLMERVPIPVKGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANR 60
EEEK+EL +L+ERVPIP++ DEP++K+N LLQ+YIS+ +L+GFA++ADMVYV QSA R
Sbjct 1069 EEEKVELAKLLERVPIPIRERLDEPAAKINALLQSYISRQRLDGFALVADMVYVTQSAGR 1128
Query 61 IMRAIFDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLPF 120
IMRAIF+I LRRGW+ +A +L C I+ R+W TM+PLRQF P E++R++EKK+ P+
Sbjct 1129 IMRAIFEISLRRGWSSVATLSLDTCKMIEKRLWPTMSPLRQFPNCPSEVIRRVEKKEFPW 1188
Query 121 DRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELAAFVQPLSRSCLVVELTITPDFQW 180
RY+DL E+GELV VPK G+ ++ ++ SFP+L + A VQP++RS + VEL I F W
Sbjct 1189 QRYFDLDPAELGELVGVPKEGRRVYNMVQSFPRLSVEAHVQPITRSLVRVELVINSQFNW 1248
Query 181 DQKIHGNGEVFWIL 194
D + G E FWIL
Sbjct 1249 DDHLSGTSEAFWIL 1262
Score = 42.7 bits (99), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 39/181 (21%), Positives = 83/181 (45%), Gaps = 12/181 (6%)
Query 3 EKLELQRLMERVPIPVKG-SPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANRI 61
E + L+R+ R+P+ + + ++P +K +LL A+ S+ +L ++ D ++ + +
Sbjct 1900 EDIVLERIHSRLPVRLSNPNYEDPHTKSFILLAAHFSRFELPP-GLVIDQKFILTRVHNL 1958
Query 62 MRAIFDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLP-- 119
+ A D G IR +++ + +W +PL+Q + L+ + K+ +
Sbjct 1959 LGACVDTLSSEGHLIACIRPMEMSQMVTQALWDRDSPLKQIPYFDDALIERCNKEGVHDV 2018
Query 120 FDRYYDLTSTEIGELVRVPKMG-KLLHRLIHSFPKLEL------AAFVQPLSRSCLVVEL 172
FD DL + EL+ + I+ +P +++ + V S S L+V+L
Sbjct 2019 FD-IIDLDDEKRTELLHMDNAHLAKCAEFINKYPDIDIDFEIEDSEDVHANSPSVLIVQL 2077
Query 173 T 173
T
Sbjct 2078 T 2078
> YER172c
Length=2163
Score = 180 bits (457), Expect = 1e-45, Method: Compositional matrix adjust.
Identities = 82/194 (42%), Positives = 127/194 (65%), Gaps = 1/194 (0%)
Query 2 EEKLELQRLMERVPIPVKGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANRI 61
EEK EL++L+E+ PIP++ D+P +KVNVLLQ+Y S+LK EGFA+ +D+V++ Q+A R+
Sbjct 1049 EEKRELKQLLEKAPIPIREDIDDPLAKVNVLLQSYFSQLKFEGFALNSDIVFIHQNAGRL 1108
Query 62 MRAIFDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLPFD 121
+RA+F+ICL+RGW L LC +MW T PLRQFK P E+++++E +P+
Sbjct 1109 LRAMFEICLKRGWGHPTRMLLNLCKSATTKMWPTNCPLRQFKTCPVEVIKRLEASTVPWG 1168
Query 122 RYYDL-TSTEIGELVRVPKMGKLLHRLIHSFPKLELAAFVQPLSRSCLVVELTITPDFQW 180
Y L T E+G +R K GK ++ L+ FPK+ + QP++RS + + I D+ W
Sbjct 1169 DYLQLETPAEVGRAIRSEKYGKQVYDLLKRFPKMSVTCNAQPITRSVMRFNIEIIADWIW 1228
Query 181 DQKIHGNGEVFWIL 194
D +HG+ E F ++
Sbjct 1229 DMNVHGSLEPFLLM 1242
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 39/157 (24%), Positives = 70/157 (44%), Gaps = 10/157 (6%)
Query 7 LQRLMERVPI--PVKGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANRIMRA 64
L +L +R+P+ P S S KV +LLQAY S+L+L D+ + + ++
Sbjct 1902 LVKLSKRLPLRFPEHTSSGSVSFKVFLLLQAYFSRLELP-VDFQNDLKDILEKVVPLINV 1960
Query 65 IFDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLPFDRYY 124
+ DI G+ A A+ L + +W PLRQ +++ K K++ + Y
Sbjct 1961 VVDILSANGYLN-ATTAMDLAQMLIQGVWDVDNPLRQIPHFNNKILEKC--KEINVETVY 2017
Query 125 DLTSTEIGE----LVRVPKMGKLLHRLIHSFPKLELA 157
D+ + E E L + ++++P +EL
Sbjct 2018 DIMALEDEERDEILTLTDSQLAQVAAFVNNYPNVELT 2054
> Hs14747366
Length=1866
Score = 147 bits (371), Expect = 1e-35, Method: Compositional matrix adjust.
Identities = 71/195 (36%), Positives = 117/195 (60%), Gaps = 2/195 (1%)
Query 1 EEEKLELQRLMER-VPIPVKGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSAN 59
EEE EL L+ + G + K+N+LLQ YIS+ +++ F++++D YV Q+A
Sbjct 692 EEEIEELDTLLSNFCELSTPGGVENSYGKINILLQTYISRGEMDSFSLISDSAYVAQNAA 751
Query 60 RIMRAIFDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLP 119
RI+RA+F+I LR+ W + R L L I R+W +PLRQF +LP ++ ++E+K L
Sbjct 752 RIVRALFEIALRKRWPTMTYRLLNLSKVIDKRLWGWASPLRQFSILPPHILTRLEEKKLT 811
Query 120 FDRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELAAFVQPLSRSCLVVELTITPDFQ 179
D+ D+ EIG ++ +G + + +H P + + A +QP++R+ L V L+I DF
Sbjct 812 VDKLKDMRKDEIGHILHHVNIGLKVKQCVHQIPSVMMEASIQPITRTVLRVTLSIYADFT 871
Query 180 WDQKIHGN-GEVFWI 193
W+ ++HG GE +WI
Sbjct 872 WNDQVHGTVGEPWWI 886
Score = 39.3 bits (90), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 23/77 (29%), Positives = 40/77 (51%), Gaps = 6/77 (7%)
Query 16 IPVKGSP---DEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANRIMRAIFDICLRR 72
+P++ +P D P +K ++LLQA++S+ L D V A R+ +A+ D+ +
Sbjct 1539 LPIESNPHSFDSPHTKAHLLLQAHLSRAMLPCPDYDTDTKTVLDQALRVCQAMLDVAANQ 1598
Query 73 GWAQLAIRALQLCNEIQ 89
GW + L + N IQ
Sbjct 1599 GW---LVTVLNITNLIQ 1612
> At5g61140
Length=2137
Score = 145 bits (365), Expect = 6e-35, Method: Compositional matrix adjust.
Identities = 75/201 (37%), Positives = 115/201 (57%), Gaps = 23/201 (11%)
Query 1 EEEKLELQRLMERV-PIPVKGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSAN 59
EEE+ EL+ L P+ VKG P K+++L+Q YIS+ ++ F++++D Y+ S
Sbjct 1058 EEEQHELETLARSCCPLEVKGGPSNKHGKISILIQLYISRGSIDAFSLVSDASYISASLA 1117
Query 60 RIMRAIFDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLP 119
RIMRA+F+ICLR+GW ++ + L+ C + ++W PLRQF ++DLP
Sbjct 1118 RIMRALFEICLRKGWCEMTLFMLEYCKAVDRQLWPHQHPLRQF------------ERDLP 1165
Query 120 FDR------YYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELAAFVQPLSRSCLVVELT 173
DR Y++ EIG L+R G R + FP ++LAA V P++R+ L V+L
Sbjct 1166 SDRRDDLDHLYEMEEKEIGALIRYNPGG----RHLGYFPSIQLAATVSPITRTVLKVDLL 1221
Query 174 ITPDFQWDQKIHGNGEVFWIL 194
ITP+F W + HG +WIL
Sbjct 1222 ITPNFIWKDRFHGTALRWWIL 1242
Score = 31.6 bits (70), Expect = 0.93, Method: Compositional matrix adjust.
Identities = 12/37 (32%), Positives = 20/37 (54%), Gaps = 0/37 (0%)
Query 58 ANRIMRAIFDICLRRGWAQLAIRALQLCNEIQGRMWS 94
AN + +A+ DIC GW ++ ++L + MWS
Sbjct 1936 ANLLFQAMIDICANSGWLSSSLTCMRLLQMVMQGMWS 1972
> SPBC13G1.10c
Length=1935
Score = 144 bits (364), Expect = 9e-35, Method: Compositional matrix adjust.
Identities = 78/197 (39%), Positives = 113/197 (57%), Gaps = 5/197 (2%)
Query 1 EEEKLELQRLMERV-PIPVKGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSAN 59
E E EL+ LME P ++ S S KVNV+LQ+YIS+ +E F + +D YV Q+A
Sbjct 837 ENEHRELESLMENSSPCQLRDSISNTSGKVNVILQSYISRAHVEDFTLTSDTNYVAQNAG 896
Query 60 RIMRAIFDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKK--D 117
RI RA+F+I + R WA A L L I R WS PL QF LP +L K+E +
Sbjct 897 RITRALFEIAMSRTWAS-AFTILSLNKSIDRRQWSFEHPLLQFD-LPHDLAVKVENQCGS 954
Query 118 LPFDRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELAAFVQPLSRSCLVVELTITPD 177
L + D+++ E+G+L+ KMG + + I P L + + PL+++ L + L ITP+
Sbjct 955 LSLEELSDMSTGELGDLIHNRKMGPTVKKFISKLPLLNINVDLLPLTKNVLRLVLNITPN 1014
Query 178 FQWDQKIHGNGEVFWIL 194
F WD + HGN ++FWI
Sbjct 1015 FNWDMRYHGNSQMFWIF 1031
> YGR271w
Length=1967
Score = 143 bits (360), Expect = 3e-34, Method: Compositional matrix adjust.
Identities = 72/195 (36%), Positives = 114/195 (58%), Gaps = 2/195 (1%)
Query 1 EEEKLELQRLM-ERVPIPVKGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSAN 59
EEE EL+RL E V + D P K NVLLQAYIS+ ++ A+ +D YV Q++
Sbjct 845 EEESKELKRLSDESVECQIGSQLDTPQGKANVLLQAYISQTRIFDSALSSDSNYVAQNSV 904
Query 60 RIMRAIFDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLP 119
RI RA+F I + R W + + L +C I+ R+W+ PL QF LPE ++R+I
Sbjct 905 RICRALFLIGVNRRWGKFSNVMLNICKSIEKRLWAFDHPLCQFD-LPENIIRRIRDTKPS 963
Query 120 FDRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELAAFVQPLSRSCLVVELTITPDFQ 179
+ +L + E+GELV K G L++++ FPK+ + A + P++ + + + + + PDF
Sbjct 964 MEHLLELEADELGELVHNKKAGSRLYKILSRFPKINIEAEIFPITTNVMRIHIALGPDFV 1023
Query 180 WDQKIHGNGEVFWIL 194
WD +IHG+ + FW+
Sbjct 1024 WDSRIHGDAQFFWVF 1038
Score = 37.4 bits (85), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 27/97 (27%), Positives = 47/97 (48%), Gaps = 5/97 (5%)
Query 24 EPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANRIMRAIFDICLRRGWAQLAIRALQ 83
+P K +LLQA++S++ L + D V V + RI++A D+ G+ + ++
Sbjct 1707 DPHVKTFLLLQAHLSRVDLPIADYIQDTVSVLDQSLRILQAYIDVASELGYFHTVLTMIK 1766
Query 84 LCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLPF 120
+ I+ W P+ VLP +R+I KD F
Sbjct 1767 MMQCIKQGYWYEDDPV---SVLPGLQLRRI--KDYTF 1798
> 7300033
Length=1809
Score = 117 bits (292), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 59/175 (33%), Positives = 97/175 (55%), Gaps = 1/175 (0%)
Query 20 GSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANRIMRAIFDICLRRGWAQLAI 79
G + KVN+L+Q Y+S ++ F++ +DM Y+ + RI RA+F I LR+ A L+
Sbjct 684 GGSENVHGKVNILIQTYLSNGYVKSFSLSSDMSYITTNIGRISRALFSIVLRQNNAVLSG 743
Query 80 RALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLPFDRYYDLTSTEIGELVRVPK 139
LQLC + R W L+QF + E + K+E++ L R D+ E+ E +R
Sbjct 744 NMLQLCKMFERRQWDFDCHLKQFPTINAETIDKLERRGLSVYRLRDMEHRELKEWLRSNT 803
Query 140 MGKLLHRLIHSFPKLELAAFVQPLSRSCLVVELTITPDFQWDQKIHGNG-EVFWI 193
L+ R H P LE+ A +QP++R+ L +++ I P F W+ ++HG + FW+
Sbjct 804 YADLVIRSAHELPLLEVEASLQPITRTVLRIKVDIWPSFTWNDRVHGKTCQSFWL 858
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 35/137 (25%), Positives = 56/137 (40%), Gaps = 9/137 (6%)
Query 1 EEEKLELQRLMERVPIPVKGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANR 60
EE E+ R P S D +K +LLQA+ ++ L + D +A R
Sbjct 1503 NEEMAEVSRFR-----PPSSSWDSSYTKTFLLLQAHFARQSLPNSDYLTDTKSALDNATR 1557
Query 61 IMRAIFDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELMRKIEK-KDLP 119
+M+A+ D RGW + QL + W + +F LP ++ ++P
Sbjct 1558 VMQAMVDYTAERGWLSTTLVVQQLMQSVIQARWFDGS---EFLTLPGVNEDNLDAFLNIP 1614
Query 120 FDRYYDLTSTEIGELVR 136
D Y LT + EL +
Sbjct 1615 HDDYDYLTLPVLKELCK 1631
> CE20305
Length=1798
Score = 103 bits (257), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 62/191 (32%), Positives = 108/191 (56%), Gaps = 7/191 (3%)
Query 1 EEEKLELQRLMER-VPIPVKGSP-DEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSA 58
EEE +L+ LM + V+G + KVNVLLQ+ IS+ A+M++ +YVQQ+A
Sbjct 658 EEEIGDLEELMSYGCMMNVRGGGLASVAGKVNVLLQSLISRTSTRNSALMSEQLYVQQNA 717
Query 59 NRIMRAIFDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQF---KVLPEELMRKIEK 115
R+ RA+F++ L+ GW+Q A L + ++ +MW LRQF +P + KIE+
Sbjct 718 GRLCRAMFEMVLKNGWSQAANAFLGIAKCVEKQMWMNQCALRQFIQIINIPITWIEKIER 777
Query 116 KDLPFDRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELAAFVQPLSRSCLVVELTIT 175
K +L++ ++G + G L+ + P++++ A +P++ + + VE+T+T
Sbjct 778 KKARESDLLELSAKDLGYMFSCD--GDRLYTYLRYLPRMDVQAKFKPITYTIVQVEVTLT 835
Query 176 PDFQWDQKIHG 186
P F W+ +IHG
Sbjct 836 PTFIWNDQIHG 846
> ECU06g0920
Length=1481
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 50/189 (26%), Positives = 87/189 (46%), Gaps = 25/189 (13%)
Query 7 LQRLMERVPIPVKGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANRIMRAIF 66
++ L VPIP + S VLLQ Y++ ++E ++ Q+ R+ RA+F
Sbjct 733 MESLRGLVPIPTE-------SSFGVLLQCYVAN-RIESTSL-------SQNLCRMFRALF 777
Query 67 DICLRRGWAQLAIRALQL--CNEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLPFDRYY 124
+I +R+ +L I + L C + R++ TPLRQF + +R +E K++PF
Sbjct 778 EIGVRK---RLGISKMILGWCKAAEHRIFPYQTPLRQF-ADDKNALRDLEMKEIPFGMLE 833
Query 125 DLTSTEIGELVRVPKMGKLLHRLIHSFPKLELAAFVQPLSRSCLVVELTITPDFQWDQKI 184
L + E V + G + H + P+ ++ V+ V+ L + F D K+
Sbjct 834 ILGKEGLDE-VGIRGSGIIEH--LRYVPRFSISPSVRVAESGYYVISLGMEKAFD-DSKV 889
Query 185 HGNGEVFWI 193
H N +I
Sbjct 890 HSNTYYLFI 898
> Hs18575318
Length=139
Score = 29.6 bits (65), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 24/88 (27%), Positives = 39/88 (44%), Gaps = 2/88 (2%)
Query 51 MVYVQQSANRIMRAIFDICLRRGWAQLAIRALQLCNEIQGRMWSTMTPLRQFKVLPEELM 110
+ VQ + +IM + R+ W + L + E+ G W + Q KVL E+ +
Sbjct 9 LTIVQATNEKIMYLATFLTARKNWNTERLSQLGITGEV-GAGW-ILRDQVQVKVLREKRL 66
Query 111 RKIEKKDLPFDRYYDLTSTEIGELVRVP 138
RK D F + L++T + L VP
Sbjct 67 RKQRTVDELFQSWLALSATSVSLLDAVP 94
> ECU07g0830
Length=1058
Score = 28.9 bits (63), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 21/89 (23%), Positives = 46/89 (51%), Gaps = 3/89 (3%)
Query 1 EEEKLELQRLMERVPIPVKGSPDEPSSKVNVLLQAYISKLKLEGFAMMADMVYVQQSANR 60
+E++ ++ L E + I + S + K+ L++A+I + + F++M D YV ++ R
Sbjct 622 DEDEESIKELCEDLGIKYEVSVE---CKLMALVKAHIKRHPVTRFSLMCDGEYVIKNLRR 678
Query 61 IMRAIFDICLRRGWAQLAIRALQLCNEIQ 89
I+ + + + +G L +R L I+
Sbjct 679 ILMGLCQVLMFQGTHLLVVRCSILLKRIE 707
> At5g05890
Length=455
Score = 28.9 bits (63), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 25/102 (24%), Positives = 42/102 (41%), Gaps = 27/102 (26%)
Query 86 NEIQGRMWSTMTPLRQFKVLPEELMRKIEKKDLPFDRYYDLTSTEIGELVR-VPKMGKLL 144
++GR W + +PEE+M K+ +K G++V+ P+ L
Sbjct 307 GSVRGREW--------IETIPEEIMEKLNEK---------------GKIVKWAPQQDVLK 343
Query 145 HRLIHSFPKLELAAFVQPLSRSCLVVELTITPDFQWDQKIHG 186
HR I F L + + C V + P F+WDQ ++
Sbjct 344 HRAIGGF--LTHNGWSSTVESVCEAVPMICLP-FRWDQMLNA 382
Lambda K H
0.324 0.139 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3264145066
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40