bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0841_orf1
Length=217
Score E
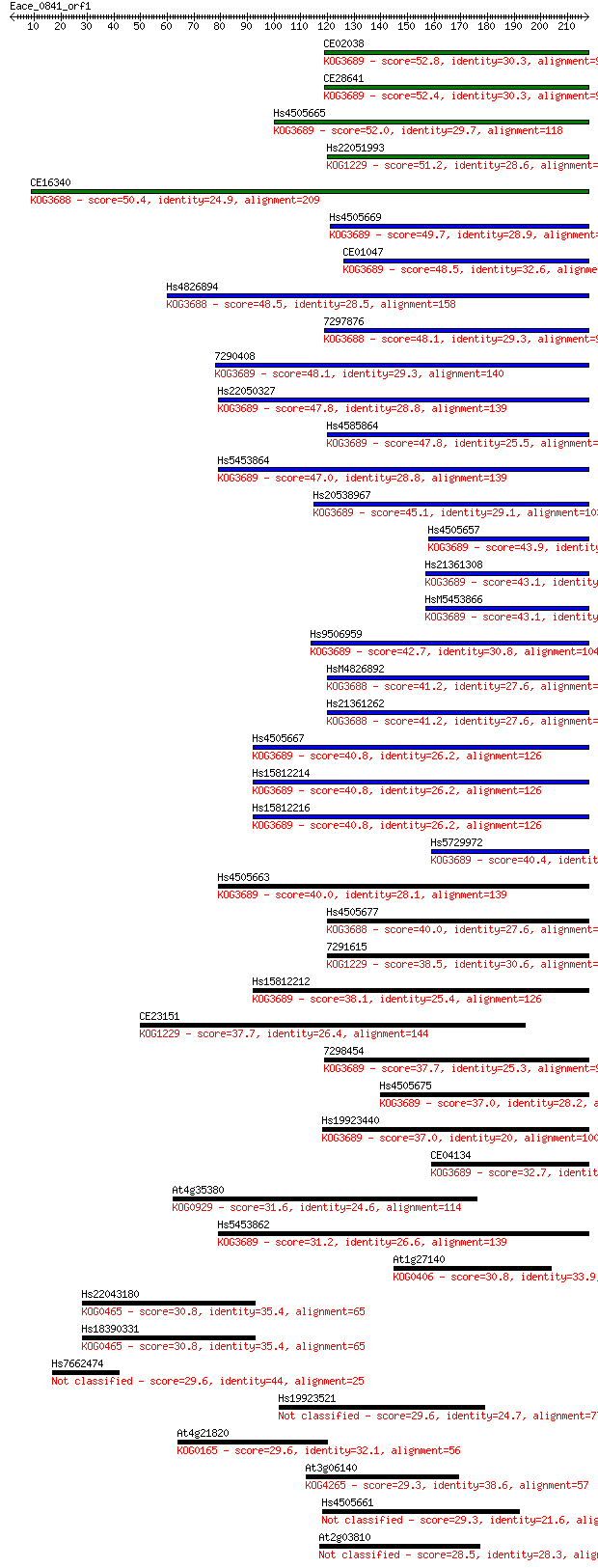
Sequences producing significant alignments: (Bits) Value
CE02038 52.8 5e-07
CE28641 52.4 7e-07
Hs4505665 52.0 9e-07
Hs22051993 51.2 2e-06
CE16340 50.4 2e-06
Hs4505669 49.7 5e-06
CE01047 48.5 9e-06
Hs4826894 48.5 1e-05
7297876 48.1 1e-05
7290408 48.1 1e-05
Hs22050327 47.8 2e-05
Hs4585864 47.8 2e-05
Hs5453864 47.0 3e-05
Hs20538967 45.1 1e-04
Hs4505657 43.9 3e-04
Hs21361308 43.1 5e-04
HsM5453866 43.1 5e-04
Hs9506959 42.7 5e-04
HsM4826892 41.2 0.002
Hs21361262 41.2 0.002
Hs4505667 40.8 0.002
Hs15812214 40.8 0.002
Hs15812216 40.8 0.002
Hs5729972 40.4 0.002
Hs4505663 40.0 0.003
Hs4505677 40.0 0.004
7291615 38.5 0.011
Hs15812212 38.1 0.013
CE23151 37.7 0.016
7298454 37.7 0.018
Hs4505675 37.0 0.028
Hs19923440 37.0 0.029
CE04134 32.7 0.54
At4g35380 31.6 1.3
Hs5453862 31.2 1.8
At1g27140 30.8 2.3
Hs22043180 30.8 2.4
Hs18390331 30.8 2.4
Hs7662474 29.6 4.4
Hs19923521 29.6 4.9
At4g21820 29.6 5.2
At3g06140 29.3 5.7
Hs4505661 29.3 6.8
At2g03810 28.5 9.5
> CE02038
Length=549
Score = 52.8 bits (125), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 30/100 (30%), Positives = 46/100 (46%), Gaps = 1/100 (1%)
Query 119 DWSFDCLHFGEISRK-PLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKENPYHN 177
DW D E+S+ L + F++L +L +H+ L +L + Y+ N YHN
Sbjct 224 DWGPDVFKIDELSKNHSLTVVTFSLLRQRNLFKTFEIHQSTLVTYLLNLEHHYRNNHYHN 283
Query 178 ALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
+H A V LL + E +DLE ++A A HD
Sbjct 284 FIHAADVAQSMHVLLMSPVLTEVFTDLEVLAAIFAGAVHD 323
> CE28641
Length=626
Score = 52.4 bits (124), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 30/100 (30%), Positives = 46/100 (46%), Gaps = 1/100 (1%)
Query 119 DWSFDCLHFGEISRK-PLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKENPYHN 177
DW D E+S+ L + F++L +L +H+ L +L + Y+ N YHN
Sbjct 301 DWGPDVFKIDELSKNHSLTVVTFSLLRQRNLFKTFEIHQSTLVTYLLNLEHHYRNNHYHN 360
Query 178 ALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
+H A V LL + E +DLE ++A A HD
Sbjct 361 FIHAADVAQSMHVLLMSPVLTEVFTDLEVLAAIFAGAVHD 400
> Hs4505665
Length=712
Score = 52.0 bits (123), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 35/120 (29%), Positives = 49/120 (40%), Gaps = 2/120 (1%)
Query 100 FGVVESLSAMCEQSAKIGTDWSFDCLHFGEIS-RKPLCEIGFAILSPFSLSPEISLHRDV 158
FGV + + W D ++S +PL I F+I L + D
Sbjct 310 FGVQTDQEEQLAKELEDTNKWGLDVFKVADVSGNRPLTAIIFSIFQERDLLKTFQIPADT 369
Query 159 LTAFLEEVTDCYKEN-PYHNALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
L +L + Y N YHN+LH A V T LL + +DLE ++A AS HD
Sbjct 370 LATYLLMLEGHYHANVAYHNSLHAADVAQSTHVLLATPALEAVFTDLEILAALFASAIHD 429
> Hs22051993
Length=761
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 28/100 (28%), Positives = 48/100 (48%), Gaps = 2/100 (2%)
Query 120 WSFDCLHFGEISRK-PLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCY-KENPYHN 177
W F+ I+ K PL +G + S F + ++ L A+ + + Y N YHN
Sbjct 433 WDFNIFELEAITHKRPLVYLGLKVFSRFGVCEFLNCSETTLRAWFQVIEANYHSSNAYHN 492
Query 178 ALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
+ H A V H T L +++ + L++++A IA+ HD
Sbjct 493 STHAADVLHATAFFLGKERVKGSLDQLDEVAALIAATVHD 532
> CE16340
Length=664
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 52/226 (23%), Positives = 90/226 (39%), Gaps = 34/226 (15%)
Query 9 PLECVIRNLDLIQRILFNANMEE-----DEREQVQELLNEARHNLTFTDNIYRFNADTFS 63
PLE + RN++ +L A M+E DE + + E+ E T D + + A TF+
Sbjct 166 PLEDLKRNIEYAALVLETAYMDETRRICDEDDDLAEVTPE-----TVPDEVREWLAATFT 220
Query 64 DGFARSYVHDLRREGISRPRKLSRMTSLAARRKSRLF--------GVVE-SLSAMCEQSA 114
R+ + R + S+A ++ +F VV+ + +
Sbjct 221 ------------RQNAGKKRDKPKFKSVANAIRTGIFFEKLFRKQQVVQCPIPPEIAELM 268
Query 115 KIGTDWSFDCLHFGEISRK-PLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKE- 172
K WSF E+S L +GF + + + + L +L + Y +
Sbjct 269 KEVCTWSFSPFQLNEVSEGHALKYVGFELFNRYGFMDRFKVPLTALENYLSALEVGYSKH 328
Query 173 -NPYHNALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
NPYHN +H A V + +L + + DLE ++ +L HD
Sbjct 329 NNPYHNVVHAADVTQSSHFMLSQTGLANSLGDLELLAVLFGALIHD 374
> Hs4505669
Length=854
Score = 49.7 bits (117), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 28/101 (27%), Positives = 47/101 (46%), Gaps = 4/101 (3%)
Query 121 SFDC--LHFGEI--SRKPLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKENPYH 176
+FD HF ++ + L + G + + + + ++VL FL ++ Y+ YH
Sbjct 498 TFDIYEFHFSDLECTELDLVKCGIQMYYELGVVRKFQIPQEVLVRFLFSISKGYRRITYH 557
Query 177 NALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
N HG V LL +++ Y +DLE + A LCHD
Sbjct 558 NWRHGFNVAQTMFTLLMTGKLKSYYTDLEAFAMVTAGLCHD 598
> CE01047
Length=918
Score = 48.5 bits (114), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 30/92 (32%), Positives = 43/92 (46%), Gaps = 8/92 (8%)
Query 126 HFGEISRKPLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKENPYHNALHGATVC 185
HF S ++GF++L L++ L+ + V+ Y+ PYHN H V
Sbjct 439 HFHRASMMFFEDLGFSML--------YKLNKRKLSYLVLRVSAGYRPVPYHNWSHAFAVT 490
Query 186 HMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
H L IR +SD+E +S IA LCHD
Sbjct 491 HFCWLTLRTDAIRRALSDMERLSLLIACLCHD 522
> Hs4826894
Length=634
Score = 48.5 bits (114), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 45/169 (26%), Positives = 71/169 (42%), Gaps = 14/169 (8%)
Query 60 DTFSDGFARSYVHDLRREGISRPRKLSRMTSLAA--------RRKSRLFGVVESLSAMCE 111
D + F R LRR +PR S + ++ A RR S + G+ S
Sbjct 104 DWLASTFTRQMGMMLRRSD-EKPRFKSIVHAVQAGIFVERMYRRTSNMVGL--SYPPAVI 160
Query 112 QSAKIGTDWSFDCLHFGEISRK-PLCEIGFAILSPFSLSPEISLHRDVLTAFLE--EVTD 168
++ K WSFD E S L I + +L+ + L + L +F+E EV
Sbjct 161 EALKDVDKWSFDVFSLNEASGDHALKFIFYELLTRYDLISRFKIPISALVSFVEALEVGY 220
Query 169 CYKENPYHNALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
+NPYHN +H A V LL + ++++LE + ++ HD
Sbjct 221 SKHKNPYHNLMHAADVTQTVHYLLYKTGVANWLTELEIFAIIFSAAIHD 269
> 7297876
Length=605
Score = 48.1 bits (113), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 29/105 (27%), Positives = 51/105 (48%), Gaps = 9/105 (8%)
Query 119 DWSFDCLHFGEISRKPLCE-IGFAILSPFSLSPEISLHRDVLTAFLEEVTD--CYKENPY 175
DW+FD E + + + + + + + + + + +L AFL V + C NPY
Sbjct 180 DWTFDVFALTEAASGQVVKYVAYELFNRYGSIHKFKIAPGILEAFLHRVEEGYCRYRNPY 239
Query 176 HNALHGATV---CHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
HN LH V H LC ++ +++DLE ++ +A+L HD
Sbjct 240 HNNLHAVDVMQTIHYCLCNTGLMN---WLTDLEIFASLLAALLHD 281
> 7290408
Length=624
Score = 48.1 bits (113), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 41/148 (27%), Positives = 58/148 (39%), Gaps = 17/148 (11%)
Query 78 GISRPRKLSRMTSLAARRKSRLFGVVESLSAMCEQSAKIGT------DWSFDCLHFGEIS 131
G+ RP LS S R FGV + ++GT W GE S
Sbjct 189 GVKRP--LSHTNSFTGERLPT-FGV------ETPRENELGTLLGELDTWGIQIFSIGEFS 239
Query 132 -RKPLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCY-KENPYHNALHGATVCHMTL 189
+PL + + I L + + F+ + D Y K+NP+HN+LH A V T
Sbjct 240 VNRPLTCVAYTIFQSRELLTSLMIPPKTFLNFMSTLEDHYVKDNPFHNSLHAADVTQSTN 299
Query 190 CLLEMLQIREYMSDLEDISACIASLCHD 217
LL + + LE A A+ HD
Sbjct 300 VLLNTPALEGVFTPLEVGGALFAACIHD 327
> Hs22050327
Length=641
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 40/141 (28%), Positives = 56/141 (39%), Gaps = 3/141 (2%)
Query 79 ISRPRKLSRMTSLAARRKSRLFGVVESLSAMCEQSAKIGTDWSFDCLHFGEIS-RKPLCE 137
IS +KL +SL R FGV + + + W E+S +PL
Sbjct 196 ISGVKKLMHSSSLTNSSIPR-FGVKTEQEDVLAKELEDVNKWGLHVFRIAELSGNRPLTV 254
Query 138 IGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKEN-PYHNALHGATVCHMTLCLLEMLQ 196
I I L + D L +L + D Y + YHN +H A V T LL
Sbjct 255 IMHTIFQERDLLKTFKIPVDTLITYLMTLEDHYHADVAYHNNIHAADVVQSTHVLLSTPA 314
Query 197 IREYMSDLEDISACIASLCHD 217
+ +DLE ++A AS HD
Sbjct 315 LEAVFTDLEILAAIFASAIHD 335
> Hs4585864
Length=860
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 25/103 (24%), Positives = 48/103 (46%), Gaps = 8/103 (7%)
Query 120 WSFDCLHFGEISRKPLCEI-----GFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKENP 174
+ + HF ++ PL E+ G + + + + ++ L F+ ++ Y++
Sbjct 501 YEINKFHFSDL---PLTELELVKCGIQMYYELKVVDKFHIPQEALVRFMYSLSKGYRKIT 557
Query 175 YHNALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
YHN HG V LL +++ Y +DLE ++ A+ CHD
Sbjct 558 YHNWRHGFNVGQTMFSLLVTGKLKRYFTDLEALAMVTAAFCHD 600
> Hs5453864
Length=809
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 40/141 (28%), Positives = 56/141 (39%), Gaps = 3/141 (2%)
Query 79 ISRPRKLSRMTSLAARRKSRLFGVVESLSAMCEQSAKIGTDWSFDCLHFGEIS-RKPLCE 137
IS +KL +SL R FGV + + + W E+S +PL
Sbjct 364 ISGVKKLMHSSSLTNSSIPR-FGVKTEQEDVLAKELEDVNKWGLHVFRIAELSGNRPLTV 422
Query 138 IGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKEN-PYHNALHGATVCHMTLCLLEMLQ 196
I I L + D L +L + D Y + YHN +H A V T LL
Sbjct 423 IMHTIFQERDLLKTFKIPVDTLITYLMTLEDHYHADVAYHNNIHAADVVQSTHVLLSTPA 482
Query 197 IREYMSDLEDISACIASLCHD 217
+ +DLE ++A AS HD
Sbjct 483 LEAVFTDLEILAAIFASAIHD 503
> Hs20538967
Length=418
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 30/105 (28%), Positives = 50/105 (47%), Gaps = 3/105 (2%)
Query 115 KIGTDWSFDCLHFGEISR-KPLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVT-DCYKE 172
K+G +W+FD F ++ L + F + S L L L FL + D + +
Sbjct 86 KVG-NWNFDIFLFDRLTNGNSLVSLTFHLFSLHGLIEYFHLDMMKLRRFLVMIQEDYHSQ 144
Query 173 NPYHNALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
NPYHNA+H A V C L+ ++ ++ + + + IA+ HD
Sbjct 145 NPYHNAVHAADVTQAMHCYLKEPKLANSVTPWDILLSLIAAATHD 189
> Hs4505657
Length=941
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 20/60 (33%), Positives = 32/60 (53%), Gaps = 0/60 (0%)
Query 158 VLTAFLEEVTDCYKENPYHNALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
L F V Y++ PYHN +H +V H L + L++ Y+ D+E + I+ +CHD
Sbjct 638 TLARFCLMVKKGYRDPPYHNWMHAFSVSHFCYLLYKNLELTNYLEDIEIFALFISCMCHD 697
> Hs21361308
Length=858
Score = 43.1 bits (100), Expect = 5e-04, Method: Composition-based stats.
Identities = 21/61 (34%), Positives = 32/61 (52%), Gaps = 0/61 (0%)
Query 157 DVLTAFLEEVTDCYKENPYHNALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCH 216
+VLT ++ V Y+ YHN HG V LL ++++Y +DLE + A+ CH
Sbjct 543 EVLTRWMYTVRKGYRAVTYHNWQHGFNVGQTMFTLLMTGRLKKYYTDLEAFAMLAAAFCH 602
Query 217 D 217
D
Sbjct 603 D 603
> HsM5453866
Length=858
Score = 43.1 bits (100), Expect = 5e-04, Method: Composition-based stats.
Identities = 21/61 (34%), Positives = 32/61 (52%), Gaps = 0/61 (0%)
Query 157 DVLTAFLEEVTDCYKENPYHNALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCH 216
+VLT ++ V Y+ YHN HG V LL ++++Y +DLE + A+ CH
Sbjct 543 EVLTRWMYTVRKGYRAVTYHNWRHGFNVGQTMFTLLMTGRLKKYYTDLEAFAMLAAAFCH 602
Query 217 D 217
D
Sbjct 603 D 603
> Hs9506959
Length=450
Score = 42.7 bits (99), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 32/112 (28%), Positives = 54/112 (48%), Gaps = 15/112 (13%)
Query 114 AKIGTDWSFDCLHFGEISRKP-----LCEI--GFAILSPFSLSPEISLHRDVLTAFLEEV 166
+K+G W FD F ++ LC + ++ F L ++LHR FL V
Sbjct 110 SKVGM-WDFDIFLFDRLTNGNSLVTLLCHLFNTHGLIHHFKLD-MVTLHR-----FLVMV 162
Query 167 T-DCYKENPYHNALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
D + +NPYHNA+H A V C L+ ++ +++ L+ + +A+ HD
Sbjct 163 QEDYHSQNPYHNAVHAADVTQAMHCYLKEPKLASFLTPLDIMLGLLAAAAHD 214
> HsM4826892
Length=535
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 27/101 (26%), Positives = 45/101 (44%), Gaps = 3/101 (2%)
Query 120 WSFDCLHFGEISRK-PLCEIGFAILSPFSLSPEISLHRDVLTAFLE--EVTDCYKENPYH 176
WSFD E S + L + + + + + L + L F E EV +NPYH
Sbjct 160 WSFDVFALNEASGEHSLKFMIYELFTRYDLINRFKIPVSCLITFAEALEVGYSKYKNPYH 219
Query 177 NALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
N +H A V ++ I ++++LE ++ A+ HD
Sbjct 220 NLIHAADVTQTVHYIMLHTGIMHWLTELEILAMVFAAAIHD 260
> Hs21361262
Length=545
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 27/101 (26%), Positives = 45/101 (44%), Gaps = 3/101 (2%)
Query 120 WSFDCLHFGEISRK-PLCEIGFAILSPFSLSPEISLHRDVLTAFLE--EVTDCYKENPYH 176
WSFD E S + L + + + + + L + L F E EV +NPYH
Sbjct 160 WSFDVFALNEASGEHSLKFMIYELFTRYDLINRFKIPVSCLITFAEALEVGYSKYKNPYH 219
Query 177 NALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
N +H A V ++ I ++++LE ++ A+ HD
Sbjct 220 NLIHAADVTQTVHYIMLHTGIMHWLTELEILAMVFAAAIHD 260
> Hs4505667
Length=875
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 33/127 (25%), Positives = 58/127 (45%), Gaps = 5/127 (3%)
Query 92 AARRKSRLFGVVESLSAMCEQSAKIGTDWSFDCLHFGEISRKPLCEIGFAILSPFSLSPE 151
AA ++R + + Q+ KI TD+SF ++ LC I + + +L
Sbjct 532 AAEEETRELQSLAAAVVPSAQTLKI-TDFSFSDFELSDL-ETALCTI--RMFTDLNLVQN 587
Query 152 ISLHRDVLTAFLEEVTDCYKEN-PYHNALHGATVCHMTLCLLEMLQIREYMSDLEDISAC 210
+ +VL ++ V Y++N YHN H L+ +I+ ++DLE ++
Sbjct 588 FQMKHEVLCRWILSVKKNYRKNVAYHNWRHAFNTAQCMFAALKAGKIQNKLTDLEILALL 647
Query 211 IASLCHD 217
IA+L HD
Sbjct 648 IAALSHD 654
> Hs15812214
Length=865
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 33/127 (25%), Positives = 58/127 (45%), Gaps = 5/127 (3%)
Query 92 AARRKSRLFGVVESLSAMCEQSAKIGTDWSFDCLHFGEISRKPLCEIGFAILSPFSLSPE 151
AA ++R + + Q+ KI TD+SF ++ LC I + + +L
Sbjct 522 AAEEETRELQSLAAAVVPSAQTLKI-TDFSFSDFELSDL-ETALCTI--RMFTDLNLVQN 577
Query 152 ISLHRDVLTAFLEEVTDCYKEN-PYHNALHGATVCHMTLCLLEMLQIREYMSDLEDISAC 210
+ +VL ++ V Y++N YHN H L+ +I+ ++DLE ++
Sbjct 578 FQMKHEVLCRWILSVKKNYRKNVAYHNWRHAFNTAQCMFAALKAGKIQNKLTDLEILALL 637
Query 211 IASLCHD 217
IA+L HD
Sbjct 638 IAALSHD 644
> Hs15812216
Length=823
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 33/127 (25%), Positives = 58/127 (45%), Gaps = 5/127 (3%)
Query 92 AARRKSRLFGVVESLSAMCEQSAKIGTDWSFDCLHFGEISRKPLCEIGFAILSPFSLSPE 151
AA ++R + + Q+ KI TD+SF ++ LC I + + +L
Sbjct 480 AAEEETRELQSLAAAVVPSAQTLKI-TDFSFSDFELSDL-ETALCTI--RMFTDLNLVQN 535
Query 152 ISLHRDVLTAFLEEVTDCYKEN-PYHNALHGATVCHMTLCLLEMLQIREYMSDLEDISAC 210
+ +VL ++ V Y++N YHN H L+ +I+ ++DLE ++
Sbjct 536 FQMKHEVLCRWILSVKKNYRKNVAYHNWRHAFNTAQCMFAALKAGKIQNKLTDLEILALL 595
Query 211 IASLCHD 217
IA+L HD
Sbjct 596 IAALSHD 602
> Hs5729972
Length=779
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 24/60 (40%), Positives = 30/60 (50%), Gaps = 4/60 (6%)
Query 159 LTAFLEEVTDCYKENPYHNALHGATVCHMTLCLLEMLQIREYM-SDLEDISACIASLCHD 217
L F+ V Y+ PYHN H TV H C+ +LQ + +DLE IA LCHD
Sbjct 498 LCRFIMSVKKNYRRVPYHNWKHAVTVAH---CMYAILQNNHTLFTDLERKGLLIACLCHD 554
> Hs4505663
Length=564
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 39/141 (27%), Positives = 58/141 (41%), Gaps = 3/141 (2%)
Query 79 ISRPRKLSRMTSLAARRKSRLFGVVESLSAMCEQSAKIGTDWSFDCLHFGEIS-RKPLCE 137
IS +KL +SL SR FGV + + W + + S +PL
Sbjct 136 ISGVKKLMHSSSLNNTSISR-FGVNTENEDHLAKELEDLNKWGLNIFNVAGYSHNRPLTC 194
Query 138 IGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKEN-PYHNALHGATVCHMTLCLLEMLQ 196
I +AI L + D ++ + D Y + YHN+LH A V T LL
Sbjct 195 IMYAIFQERDLLKTFRISSDTFITYMMTLEDHYHSDVAYHNSLHAADVAQSTHVLLSTPA 254
Query 197 IREYMSDLEDISACIASLCHD 217
+ +DLE ++A A+ HD
Sbjct 255 LDAVFTDLEILAAIFAAAIHD 275
> Hs4505677
Length=536
Score = 40.0 bits (92), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 27/101 (26%), Positives = 45/101 (44%), Gaps = 3/101 (2%)
Query 120 WSFDCLHFGEISRK-PLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKE--NPYH 176
W FD + + L I F +L+ +L + L +FL+ + Y + NPYH
Sbjct 164 WCFDVFSLNQAADDHALRTIVFELLTRHNLISRFKIPTVFLMSFLDALETGYGKYKNPYH 223
Query 177 NALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
N +H A V C L + +S++E ++ A+ HD
Sbjct 224 NQIHAADVTQTVHCFLLRTGMVHCLSEIELLAIIFAAAIHD 264
> 7291615
Length=1060
Score = 38.5 bits (88), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 30/102 (29%), Positives = 48/102 (47%), Gaps = 4/102 (3%)
Query 120 WSFDCLHFGEISR-KPLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCY-KENPYHN 177
W FD EI+ PL +G + F + +++ +V A+L + Y K N YHN
Sbjct 728 WDFDIFKLEEITDYHPLLYLGMEMFRRFDVFATLNIDENVCKAWLAVIEAHYRKSNTYHN 787
Query 178 ALHGATVCHMTLCLLEMLQIRE--YMSDLEDISACIASLCHD 217
+ H A V T + L ++ M +E+ +A IA+ HD
Sbjct 788 STHAADVMQATGAFITQLTNKDMLVMDRMEEATALIAAAAHD 829
> Hs15812212
Length=833
Score = 38.1 bits (87), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 32/127 (25%), Positives = 57/127 (44%), Gaps = 5/127 (3%)
Query 92 AARRKSRLFGVVESLSAMCEQSAKIGTDWSFDCLHFGEISRKPLCEIGFAILSPFSLSPE 151
AA ++R + + Q+ KI TD+SF ++ LC I + + +L
Sbjct 490 AAEEETRELQSLAAAVVPSAQTLKI-TDFSFSDFELSDL-ETALCTI--RMFTDLNLVQN 545
Query 152 ISLHRDVLTAFLEEVTDCYKEN-PYHNALHGATVCHMTLCLLEMLQIREYMSDLEDISAC 210
+ +VL ++ V Y++N YHN H L+ +I+ ++DL ++
Sbjct 546 FQMKHEVLCRWILSVKKNYRKNVAYHNWRHAFNTAQCMFAALKAGKIQNKLTDLGILALL 605
Query 211 IASLCHD 217
IA+L HD
Sbjct 606 IAALSHD 612
> CE23151
Length=760
Score = 37.7 bits (86), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 38/147 (25%), Positives = 60/147 (40%), Gaps = 8/147 (5%)
Query 50 FTDNIYRFNADTFSDGFARSYVHDLRREGISRP-RKLSRMTSLAARRKSRLFGVVESLSA 108
+ +I RF +D A Y L R + P R+ R A R K G +SA
Sbjct 378 YAPSINRFRD---ADRIATQYYDGLIR--LHHPARQRKRSVVDAHREKRGSHGERRRVSA 432
Query 109 MCEQSAKIGTDWSFDCLHFGEIS-RKPLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVT 167
+ + + W FD LH ++S L ++G + + + + D+L ++ +
Sbjct 433 DVKNALENDNCWKFDILHLEKVSDHHALSQVGMKVFERWKVCDVLGCSDDLLHRWILSIE 492
Query 168 DCYKE-NPYHNALHGATVCHMTLCLLE 193
Y N YHNA H A V T L+
Sbjct 493 AHYHAGNTYHNATHAADVLQATSFFLD 519
> 7298454
Length=1284
Score = 37.7 bits (86), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 25/99 (25%), Positives = 39/99 (39%), Gaps = 3/99 (3%)
Query 119 DWSFDCLHFGEISRKPLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKENPYHNA 178
D+ FD +HF + C + + +VL +L V Y+ YHN
Sbjct 639 DFKFDDIHFEDDDTLKAC---LRMFLDLDFVERFHIDYEVLCRWLLSVKKNYRNVTYHNW 695
Query 179 LHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
H V M +L Q + ++E ++ I LCHD
Sbjct 696 RHAFNVAQMMFAILTTTQWWKIFGEIECLALIIGCLCHD 734
> Hs4505675
Length=593
Score = 37.0 bits (84), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 22/78 (28%), Positives = 40/78 (51%), Gaps = 6/78 (7%)
Query 140 FAILSPFSLSPEISLHRDVLTAFLEEVTDCYKENPYHNALHGATVCHMTLCLLEMLQIRE 199
++ FS++P ++L R +L V D Y+ NP+HN H V M ++ + ++E
Sbjct 282 LGLVRDFSINP-VTLRR-----WLFCVHDNYRNNPFHNFRHCFCVAQMMYSMVWLCSLQE 335
Query 200 YMSDLEDISACIASLCHD 217
S + + A++CHD
Sbjct 336 KFSQTDILILMTAAICHD 353
> Hs19923440
Length=934
Score = 37.0 bits (84), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 20/102 (19%), Positives = 44/102 (43%), Gaps = 2/102 (1%)
Query 118 TDWSFDCLHFGEIS--RKPLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKENPY 175
++ + D +HF + S + + + + + + L +L V Y+ Y
Sbjct 604 SELAIDDIHFDDFSLDVDAMITAALRMFMELGMVQKFKIDYETLCRWLLTVRKNYRMVLY 663
Query 176 HNALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
HN H VC + +L ++ ++++E ++ + LCHD
Sbjct 664 HNWRHAFNVCQLMFAMLTTAGFQDILTEVEILAVIVGCLCHD 705
> CE04134
Length=393
Score = 32.7 bits (73), Expect = 0.54, Method: Compositional matrix adjust.
Identities = 18/59 (30%), Positives = 28/59 (47%), Gaps = 2/59 (3%)
Query 159 LTAFLEEVTDCYKENPYHNALHGATVCHMTLCLLEMLQIREYMSDLEDISACIASLCHD 217
L F+ V Y+ YHN HG +V H L + + + LE ++ ++ LCHD
Sbjct 173 LIRFVLTVRKNYRRVAYHNWAHGWSVAHAMFATL--MNSPDAFTKLEALALYVSCLCHD 229
> At4g35380
Length=1711
Score = 31.6 bits (70), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 28/114 (24%), Positives = 45/114 (39%), Gaps = 18/114 (15%)
Query 62 FSDGFARSYVHDLRREGISRPRKLSRMTSLAARRKSRLFGVVESLSAMCEQSAKIGTDWS 121
+DG ARS D RE ++ + +T + R +E + + QS IG D
Sbjct 1428 LADGLARSASEDEWREIFLALKEAASLTFAGFMKVLRTMDDIEDVETLSGQSVNIG-DLD 1486
Query 122 FDCLHFGEISRKPLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKENPY 175
D LH + + + H DVL+ +E V+D Y+ N +
Sbjct 1487 DDSLHI----------MSYVV-------SRTKKHIDVLSQIVEVVSDLYRRNQF 1523
> Hs5453862
Length=647
Score = 31.2 bits (69), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 37/142 (26%), Positives = 57/142 (40%), Gaps = 5/142 (3%)
Query 79 ISRPRKLSRMTSLAARRKSRLFGVVESLSAMCEQSAKIGTDWSFD--CLHFGEISRKPLC 136
I+ +KL SL R FGV + Q + W + C+ R C
Sbjct 96 ITGLKKLMHSNSLNNSNIPR-FGVKTDQEELLAQELENLNKWGLNIFCVSDYAGGRSLTC 154
Query 137 EIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKEN-PYHNALHGATVCHMTLCLLEML 195
I + I L + + D + ++ + D Y + YHN+LH A V T LL
Sbjct 155 -IMYMIFQERDLLKKFRIPVDTMVTYMLTLEDHYHADVAYHNSLHAADVLQSTHVLLATP 213
Query 196 QIREYMSDLEDISACIASLCHD 217
+ +DLE ++A A+ HD
Sbjct 214 ALDAVFTDLEILAALFAAAIHD 235
> At1g27140
Length=243
Score = 30.8 bits (68), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 20/68 (29%), Positives = 31/68 (45%), Gaps = 11/68 (16%)
Query 145 PFSLSPEISLH-RDVLTAFLEEVTD--------CYKENPYHNALHGATVCHMTLCLLEML 195
PFS+ P ++LH + + +LEE D K NP H + H L + E L
Sbjct 16 PFSIRPRVALHLKSIKYEYLEEPDDDLGEKSQLLLKSNPIHKKT--PVLIHGDLAICESL 73
Query 196 QIREYMSD 203
I +Y+ +
Sbjct 74 NIVQYLDE 81
> Hs22043180
Length=485
Score = 30.8 bits (68), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 23/74 (31%), Positives = 36/74 (48%), Gaps = 13/74 (17%)
Query 28 NMEEDEREQVQELLNEARHN------LTFTDNIYRFNADTFSDGFARSYVHDLRREGI-- 79
N E+D +E+ + L+N +R N L F + RF T + RSY +L++
Sbjct 64 NKEDDSKEKTKILMNSSRDNSHPFVGLAFKLEVGRFGQLT----YVRSYQGELKKGDTIY 119
Query 80 -SRPRKLSRMTSLA 92
+R RK R+ LA
Sbjct 120 NTRTRKKVRLQRLA 133
> Hs18390331
Length=751
Score = 30.8 bits (68), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 23/74 (31%), Positives = 36/74 (48%), Gaps = 13/74 (17%)
Query 28 NMEEDEREQVQELLNEARHN------LTFTDNIYRFNADTFSDGFARSYVHDLRREGI-- 79
N E+D +E+ + L+N +R N L F + RF T + RSY +L++
Sbjct 330 NKEDDSKEKTKILMNSSRDNSHPFVGLAFKLEVGRFGQLT----YVRSYQGELKKGDTIY 385
Query 80 -SRPRKLSRMTSLA 92
+R RK R+ LA
Sbjct 386 NTRTRKKVRLQRLA 399
> Hs7662474
Length=1709
Score = 29.6 bits (65), Expect = 4.4, Method: Composition-based stats.
Identities = 11/25 (44%), Positives = 21/25 (84%), Gaps = 0/25 (0%)
Query 17 LDLIQRILFNANMEEDEREQVQELL 41
++ +Q+ L NAN+ EDE+EQ+++L+
Sbjct 1311 IEELQKNLLNANLSEDEKEQLKKLM 1335
> Hs19923521
Length=682
Score = 29.6 bits (65), Expect = 4.9, Method: Composition-based stats.
Identities = 19/77 (24%), Positives = 36/77 (46%), Gaps = 1/77 (1%)
Query 102 VVESLSAMCEQSAKIGTDWSFDCLHFGEISRKPLCEIGFAILSPFSLSPEISLHRDVLTA 161
+V+ L+ ++ A +G + C E+ + + G + P L E LHR++L
Sbjct 501 IVDILTMKSQEKANLGENMEKSCASKEEVREVSIEDTGVDV-DPEKLEMESKLHRNLLLQ 559
Query 162 FLEEVTDCYKENPYHNA 178
E+ D ++P H+A
Sbjct 560 DCEKEQDNKTKDPTHDA 576
> At4g21820
Length=1088
Score = 29.6 bits (65), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 18/56 (32%), Positives = 28/56 (50%), Gaps = 0/56 (0%)
Query 64 DGFARSYVHDLRREGISRPRKLSRMTSLAARRKSRLFGVVESLSAMCEQSAKIGTD 119
DG ARSYV++ EG+ RP + ++ +R L V++ + S K G D
Sbjct 313 DGLARSYVYNKMVEGLYRPGYYEALGNVILKRILLLVLVIDRAKSQSCLSLKYGID 368
> At3g06140
Length=546
Score = 29.3 bits (64), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 22/63 (34%), Positives = 32/63 (50%), Gaps = 8/63 (12%)
Query 112 QSAKIGTDWSFDCLHFGEISRK------PLCEIGFAILSPFSLSPEISLHRDVLTAFLEE 165
Q + GTD SF L ++S+ PL I+SP S+S + S+H+ V A LE+
Sbjct 384 QPSGTGTDLSFFVL--DDLSKPLEEDVYPLVISAETIISPNSISEQSSVHKQVTQAVLEK 441
Query 166 VTD 168
D
Sbjct 442 DND 444
> Hs4505661
Length=1112
Score = 29.3 bits (64), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 16/78 (20%), Positives = 31/78 (39%), Gaps = 4/78 (5%)
Query 118 TDWSFDCLHF----GEISRKPLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKEN 173
++W+F GE S + L ++ + + L + + + + Y++
Sbjct 675 SNWNFPIFELVEKMGEKSGRILSQVMYTLFQDTGLLEIFKIPTQQFMNYFRALENGYRDI 734
Query 174 PYHNALHGATVCHMTLCL 191
PYHN +H V H L
Sbjct 735 PYHNRIHATDVLHAVWYL 752
> At2g03810
Length=431
Score = 28.5 bits (62), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 17/60 (28%), Positives = 26/60 (43%), Gaps = 3/60 (5%)
Query 117 GTDWSFDCLHFGEISRKPLCEIGFAILSPFSLSPEISLHRDVLTAFLEEVTDCYKENPYH 176
G +W + G+ R + + SP P + ++V L E+ CYKEN YH
Sbjct 67 GEEWENEA---GKKVRDTSHDCDANVDSPEKKDPVFYMDKNVTACDLPEIVVCYKENTYH 123
Lambda K H
0.324 0.137 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4040519078
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40