bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0894_orf3
Length=98
Score E
Sequences producing significant alignments: (Bits) Value
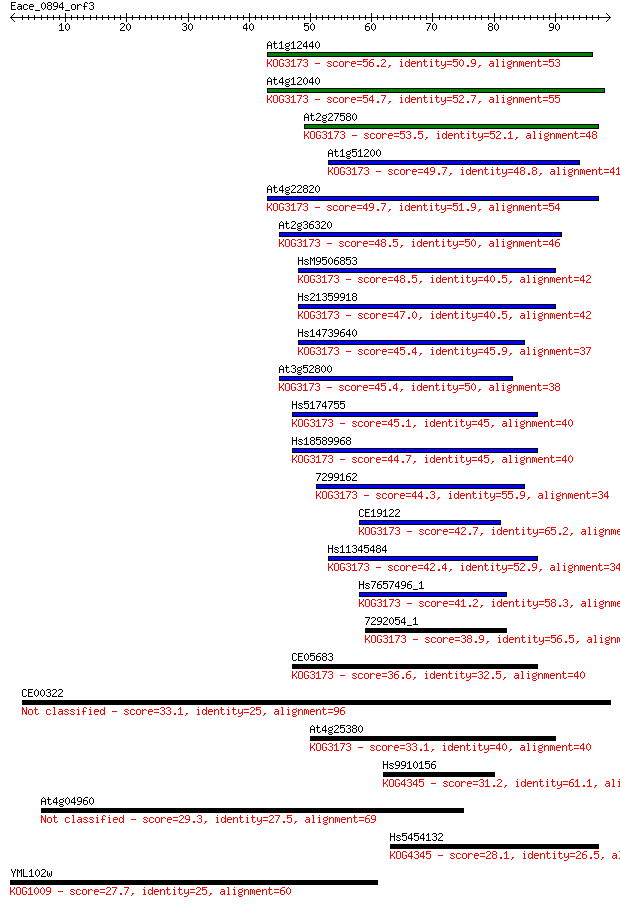
At1g12440 56.2 1e-08
At4g12040 54.7 4e-08
At2g27580 53.5 1e-07
At1g51200 49.7 1e-06
At4g22820 49.7 1e-06
At2g36320 48.5 3e-06
HsM9506853 48.5 3e-06
Hs21359918 47.0 9e-06
Hs14739640 45.4 2e-05
At3g52800 45.4 3e-05
Hs5174755 45.1 4e-05
Hs18589968 44.7 4e-05
7299162 44.3 6e-05
CE19122 42.7 2e-04
Hs11345484 42.4 2e-04
Hs7657496_1 41.2 5e-04
7292054_1 38.9 0.002
CE05683 36.6 0.011
CE00322 33.1 0.14
At4g25380 33.1 0.15
Hs9910156 31.2 0.49
At4g04960 29.3 1.7
Hs5454132 28.1 4.5
YML102w 27.7 5.5
> At1g12440
Length=168
Score = 56.2 bits (134), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 27/55 (49%), Positives = 35/55 (63%), Gaps = 2/55 (3%)
Query 43 MSSEQHENER--PSVPPLCANNCGFYGSPASRNLCSKCYREFLKAESAAAATAAA 95
M SEQ+++ PS P LC CGF+GSP++ NLCSKCYR+ E A+ AA
Sbjct 1 MGSEQNDSTSFSPSEPKLCVKGCGFFGSPSNMNLCSKCYRDIRATEEQTASAKAA 55
> At4g12040
Length=175
Score = 54.7 bits (130), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 29/57 (50%), Positives = 37/57 (64%), Gaps = 3/57 (5%)
Query 43 MSSEQHENER--PSVPPLCANNCGFYGSPASRNLCSKCYREFLKAESAAAATAAAAA 97
M SE++ + P+ P LC N CGF+GSP++ NLCSKCYR L+AE A A AA
Sbjct 1 MGSEENNSTSFPPTEPKLCDNGCGFFGSPSNMNLCSKCYRS-LRAEEDQTAVAKAAV 56
> At2g27580
Length=163
Score = 53.5 bits (127), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 25/48 (52%), Positives = 34/48 (70%), Gaps = 1/48 (2%)
Query 49 ENERPSVPPLCANNCGFYGSPASRNLCSKCYREFLKAESAAAATAAAA 96
E R P LCANNCGF+GS A++NLCSKC+R+ L+ + ++TA A
Sbjct 3 EEHRLQEPRLCANNCGFFGSTATQNLCSKCFRD-LQHQEQNSSTAKHA 49
> At1g51200
Length=173
Score = 49.7 bits (117), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 20/41 (48%), Positives = 27/41 (65%), Gaps = 0/41 (0%)
Query 53 PSVPPLCANNCGFYGSPASRNLCSKCYREFLKAESAAAATA 93
P P LC NNCGF+GS A+ N+CSKC+++ L + A A
Sbjct 12 PEGPKLCTNNCGFFGSAATMNMCSKCHKDMLFQQEQGAKFA 52
> At4g22820
Length=176
Score = 49.7 bits (117), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 28/59 (47%), Positives = 35/59 (59%), Gaps = 6/59 (10%)
Query 43 MSSEQHE-----NERPSVPPLCANNCGFYGSPASRNLCSKCYREFLKAESAAAATAAAA 96
M SEQ++ + S P LC CGF+GSP++ +LCSKCYR AE A A A AA
Sbjct 1 MGSEQNDSTSFTQSQASEPKLCVKGCGFFGSPSNMDLCSKCYRGIC-AEEAQTAVAKAA 58
> At2g36320
Length=161
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 23/47 (48%), Positives = 30/47 (63%), Gaps = 1/47 (2%)
Query 45 SEQHENERPSVPPLCANNCGFYGSPASRNLCSKCYREF-LKAESAAA 90
+E+H E P LC NNCGF+GS A+ NLCS CY + LK + A+
Sbjct 2 AEEHRCETPEGHRLCVNNCGFFGSSATMNLCSNCYGDLCLKQQQQAS 48
> HsM9506853
Length=186
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 17/42 (40%), Positives = 27/42 (64%), Gaps = 0/42 (0%)
Query 48 HENERPSVPPLCANNCGFYGSPASRNLCSKCYREFLKAESAA 89
E VP LC+ CGFYG+P + +CS CY+E L+ ++++
Sbjct 3 QETNHSQVPMLCSTGCGFYGNPRTNGMCSVCYKEHLQRQNSS 44
> Hs21359918
Length=208
Score = 47.0 bits (110), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 17/42 (40%), Positives = 27/42 (64%), Gaps = 0/42 (0%)
Query 48 HENERPSVPPLCANNCGFYGSPASRNLCSKCYREFLKAESAA 89
E VP LC+ CGFYG+P + +CS CY+E L+ ++++
Sbjct 3 QETNHSQVPMLCSTGCGFYGNPRTNGMCSVCYKEHLQRQNSS 44
> Hs14739640
Length=159
Score = 45.4 bits (106), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 17/37 (45%), Positives = 22/37 (59%), Gaps = 0/37 (0%)
Query 48 HENERPSVPPLCANNCGFYGSPASRNLCSKCYREFLK 84
E VP LC+ CGFYG+P + CS CY+E L+
Sbjct 3 QETNHSQVPMLCSTGCGFYGNPRTNGTCSVCYKEHLQ 39
> At3g52800
Length=170
Score = 45.4 bits (106), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 19/38 (50%), Positives = 24/38 (63%), Gaps = 0/38 (0%)
Query 45 SEQHENERPSVPPLCANNCGFYGSPASRNLCSKCYREF 82
+E+H + P LC NNCGF GS A+ NLCS CY +
Sbjct 2 AEEHRCQTPESNRLCVNNCGFLGSSATMNLCSNCYGDL 39
> Hs5174755
Length=213
Score = 45.1 bits (105), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 18/40 (45%), Positives = 26/40 (65%), Gaps = 1/40 (2%)
Query 47 QHENERPSVPPLCANNCGFYGSPASRNLCSKCYREFLKAE 86
Q N+ P P LC+ CGFYG+P + +CS CY+E L+ +
Sbjct 3 QETNQTPG-PMLCSTGCGFYGNPRTNGMCSVCYKEHLQRQ 41
> Hs18589968
Length=167
Score = 44.7 bits (104), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 18/40 (45%), Positives = 26/40 (65%), Gaps = 1/40 (2%)
Query 47 QHENERPSVPPLCANNCGFYGSPASRNLCSKCYREFLKAE 86
Q N+ P P LC+ CGFYG+P + +CS CY+E L+ +
Sbjct 3 QETNQTPG-PMLCSTGCGFYGNPRTNGMCSVCYKEHLQRQ 41
> 7299162
Length=97
Score = 44.3 bits (103), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 19/36 (52%), Positives = 26/36 (72%), Gaps = 2/36 (5%)
Query 51 ERPSVP--PLCANNCGFYGSPASRNLCSKCYREFLK 84
ER S P P+C + CGFYG+PA+ LCS CY++ L+
Sbjct 2 ERESNPMQPMCRSGCGFYGNPATDGLCSVCYKDSLR 37
> CE19122
Length=161
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 15/23 (65%), Positives = 17/23 (73%), Gaps = 0/23 (0%)
Query 58 LCANNCGFYGSPASRNLCSKCYR 80
LC N CGFYG+P N CSKC+R
Sbjct 50 LCVNGCGFYGTPQWENRCSKCWR 72
> Hs11345484
Length=227
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 18/34 (52%), Positives = 24/34 (70%), Gaps = 2/34 (5%)
Query 53 PSVPPLCANNCGFYGSPASRNLCSKCYREFLKAE 86
PS+PP C CGF+GS + NLCSKC+ +F K +
Sbjct 12 PSLPPRCP--CGFWGSSKTMNLCSKCFADFQKKQ 43
> Hs7657496_1
Length=60
Score = 41.2 bits (95), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 14/24 (58%), Positives = 18/24 (75%), Gaps = 0/24 (0%)
Query 58 LCANNCGFYGSPASRNLCSKCYRE 81
LC CG+YG+PA + CSKC+RE
Sbjct 18 LCKKGCGYYGNPAWQGFCSKCWRE 41
> 7292054_1
Length=175
Score = 38.9 bits (89), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 13/23 (56%), Positives = 17/23 (73%), Gaps = 0/23 (0%)
Query 59 CANNCGFYGSPASRNLCSKCYRE 81
C + CGFYG+P + LCS C+RE
Sbjct 19 CRSGCGFYGTPQNEGLCSMCFRE 41
> CE05683
Length=189
Score = 36.6 bits (83), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 13/40 (32%), Positives = 24/40 (60%), Gaps = 0/40 (0%)
Query 47 QHENERPSVPPLCANNCGFYGSPASRNLCSKCYREFLKAE 86
++E ++ P C CGF+G+ A+ CS+C++ LK +
Sbjct 2 ENEQQQAQTAPSCRAGCGFFGASATEGYCSQCFKNTLKRQ 41
> CE00322
Length=344
Score = 33.1 bits (74), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 24/96 (25%), Positives = 42/96 (43%), Gaps = 5/96 (5%)
Query 3 ALYTDTISSTSGVAAVQQTIRKSSRSSSLKFPFPLSFVVKMSSEQHENERPSVPPLCANN 62
AL+ I+ST + + + + + ++ F S +S+E H NE P+ P+ +
Sbjct 207 ALFDSVINSTREIIQIGDSGNRMPTAIESEYNFNFS-NDDISTEIHHNEEPTSKPVSSET 265
Query 63 CGFYGSPASRNLCSKCYREFLKAESAAAATAAAAAC 98
C F G+ S + C+ E+ ES A C
Sbjct 266 CKFCGNRHSTDECT----EYTTGESRRARLQKLLLC 297
> At4g25380
Length=130
Score = 33.1 bits (74), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 16/40 (40%), Positives = 23/40 (57%), Gaps = 2/40 (5%)
Query 50 NERPSVPPLCANNCGFYGSPASRNLCSKCYREFLKAESAA 89
NE ++P C CG YG+ + NLCS CY++ + S A
Sbjct 3 NETEALP--CEGGCGLYGTRVNNNLCSLCYKKSVLQHSPA 40
> Hs9910156
Length=858
Score = 31.2 bits (69), Expect = 0.49, Method: Compositional matrix adjust.
Identities = 11/18 (61%), Positives = 12/18 (66%), Gaps = 0/18 (0%)
Query 62 NCGFYGSPASRNLCSKCY 79
NC FYG P + N CS CY
Sbjct 821 NCSFYGHPETNNFCSCCY 838
> At4g04960
Length=686
Score = 29.3 bits (64), Expect = 1.7, Method: Composition-based stats.
Identities = 19/69 (27%), Positives = 33/69 (47%), Gaps = 0/69 (0%)
Query 6 TDTISSTSGVAAVQQTIRKSSRSSSLKFPFPLSFVVKMSSEQHENERPSVPPLCANNCGF 65
T+ S +G A +TIR +S PF SF+ M+ ++ + L A + G
Sbjct 51 TNQTSFATGRALYNRTIRTKDPITSSVLPFSTSFIFTMAPYKNTLPGHGIVFLFAPSTGI 110
Query 66 YGSPASRNL 74
GS ++++L
Sbjct 111 NGSSSAQHL 119
> Hs5454132
Length=790
Score = 28.1 bits (61), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 9/34 (26%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 63 CGFYGSPASRNLCSKCYREFLKAESAAAATAAAA 96
C ++G+P ++ C+ C+ E+ + + AAA+ +
Sbjct 612 CVYFGTPENKGFCTLCFIEYRENKHFAAASGKVS 645
> YML102w
Length=468
Score = 27.7 bits (60), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 15/60 (25%), Positives = 29/60 (48%), Gaps = 0/60 (0%)
Query 1 RPALYTDTISSTSGVAAVQQTIRKSSRSSSLKFPFPLSFVVKMSSEQHENERPSVPPLCA 60
RPA+ ++ + +AA ++ + S LK P+ L F + ++E + + PLC
Sbjct 308 RPAIRIPSLKKPALMAAFSPVFYETCQKSVLKLPYKLVFAIATTNEVLVYDTDVLEPLCV 367
Lambda K H
0.315 0.123 0.359
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1194657780
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40