bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0946_orf1
Length=226
Score E
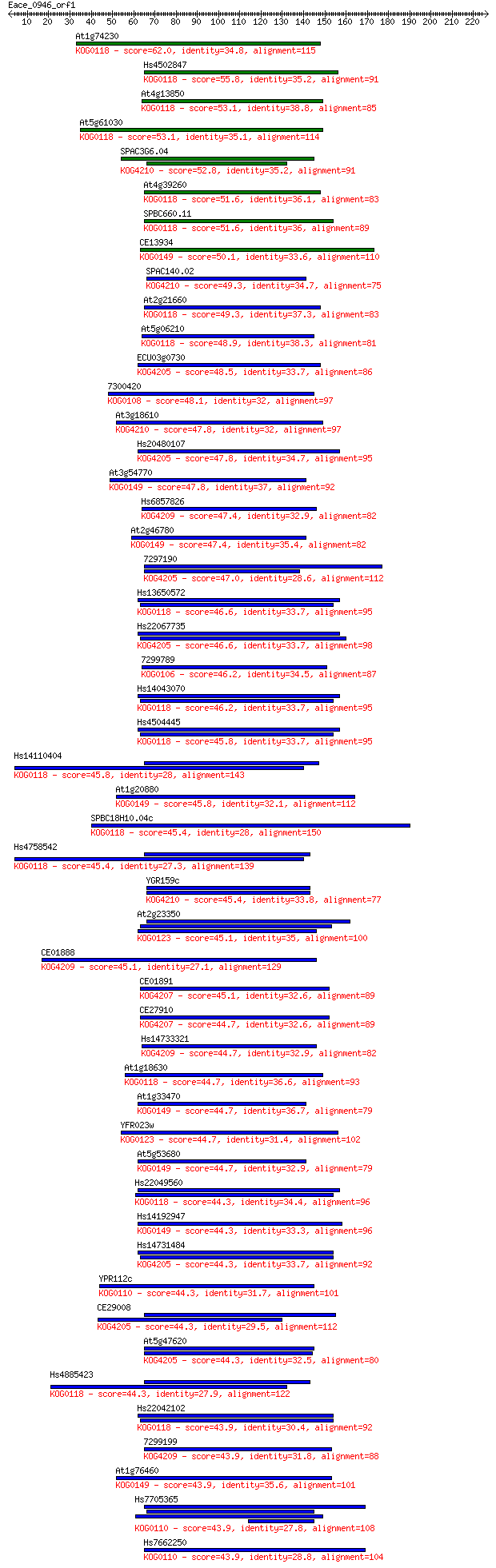
Sequences producing significant alignments: (Bits) Value
At1g74230 62.0 9e-10
Hs4502847 55.8 7e-08
At4g13850 53.1 4e-07
At5g61030 53.1 4e-07
SPAC3G6.04 52.8 6e-07
At4g39260 51.6 1e-06
SPBC660.11 51.6 1e-06
CE13934 50.1 4e-06
SPAC140.02 49.3 6e-06
At2g21660 49.3 6e-06
At5g06210 48.9 7e-06
ECU03g0730 48.5 1e-05
7300420 48.1 1e-05
At3g18610 47.8 2e-05
Hs20480107 47.8 2e-05
At3g54770 47.8 2e-05
Hs6857826 47.4 3e-05
At2g46780 47.4 3e-05
7297190 47.0 3e-05
Hs13650572 46.6 4e-05
Hs22067735 46.6 4e-05
7299789 46.2 5e-05
Hs14043070 46.2 6e-05
Hs4504445 45.8 6e-05
Hs14110404 45.8 7e-05
At1g20880 45.8 8e-05
SPBC18H10.04c 45.4 8e-05
Hs4758542 45.4 8e-05
YGR159c 45.4 1e-04
At2g23350 45.1 1e-04
CE01888 45.1 1e-04
CE01891 45.1 1e-04
CE27910 44.7 1e-04
Hs14733321 44.7 1e-04
At1g18630 44.7 1e-04
At1g33470 44.7 2e-04
YFR023w 44.7 2e-04
At5g53680 44.7 2e-04
Hs22049560 44.3 2e-04
Hs14192947 44.3 2e-04
Hs14731484 44.3 2e-04
YPR112c 44.3 2e-04
CE29008 44.3 2e-04
At5g47620 44.3 2e-04
Hs4885423 44.3 2e-04
Hs22042102 43.9 2e-04
7299199 43.9 2e-04
At1g76460 43.9 3e-04
Hs7705365 43.9 3e-04
Hs7662250 43.9 3e-04
> At1g74230
Length=289
Score = 62.0 bits (149), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 40/115 (34%), Positives = 60/115 (52%), Gaps = 8/115 (6%)
Query 33 YLQRRGRELNRELSLLAEQVNEAQGITTKSLTKVFVGGLAYSTSRYDLCELFAEVGNIKS 92
+L + GR ++ S + + Q I S +K+FVGG++YST + L E F++ G +
Sbjct 3 FLSKVGRLFSQTSSHVTASSSMLQSIRCMSSSKIFVGGISYSTDEFGLREAFSKYGEV-- 60
Query 93 IVIPQVDRSIPPDYEGAYSVRGIAFVQFEDAESAKKALQCNGKIVNGRSVVVTPA 147
VD I D E S RG AFV F E A A+Q +G+ ++GR + V A
Sbjct 61 -----VDAKIIVDRETGRS-RGFAFVTFTSTEEASNAMQLDGQDLHGRRIRVNYA 109
> Hs4502847
Length=172
Score = 55.8 bits (133), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 32/92 (34%), Positives = 51/92 (55%), Gaps = 9/92 (9%)
Query 65 KVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFEDAE 124
K+FVGGL++ T+ L ++F++ G I +V+ + DR RG FV FE+ +
Sbjct 7 KLFVGGLSFDTNEQSLEQVFSKYGQISEVVVVK-DRETQR-------SRGFGFVTFENID 58
Query 125 SAKKALQC-NGKIVNGRSVVVTPANLTREERA 155
AK A+ NGK V+GR + V A + + R+
Sbjct 59 DAKDAMMAMNGKSVDGRQIRVDQAGKSSDNRS 90
> At4g13850
Length=158
Score = 53.1 bits (126), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 33/86 (38%), Positives = 47/86 (54%), Gaps = 9/86 (10%)
Query 64 TKVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFEDA 123
TK+F+GGL++ T L + FA G++ VD + D E S RG FV F D
Sbjct 35 TKLFIGGLSWGTDDASLRDAFAHFGDV-------VDAKVIVDRETGRS-RGFGFVNFNDE 86
Query 124 ESAKKAL-QCNGKIVNGRSVVVTPAN 148
+A A+ + +GK +NGR + V PAN
Sbjct 87 GAATAAISEMDGKELNGRHIRVNPAN 112
> At5g61030
Length=309
Score = 53.1 bits (126), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 40/115 (34%), Positives = 61/115 (53%), Gaps = 11/115 (9%)
Query 35 QRRGRELNRELSLLAEQVNEAQGITTKSLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIV 94
Q ++LN ++SL + + +A I S +K+F+GG+AYS L E F + G +
Sbjct 13 QTTNKQLNAQVSLSSPSLFQA--IRCMSSSKLFIGGMAYSMDEDSLREAFTKYGEV---- 66
Query 95 IPQVDRSIPPDYEGAYSVRGIAFVQFEDAESAKKALQC-NGKIVNGRSVVVTPAN 148
VD + D E S RG FV F +E+A A+Q +G+ ++GR V V AN
Sbjct 67 ---VDTRVILDRETGRS-RGFGFVTFTSSEAASSAIQALDGRDLHGRVVKVNYAN 117
> SPAC3G6.04
Length=369
Score = 52.8 bits (125), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 32/102 (31%), Positives = 55/102 (53%), Gaps = 11/102 (10%)
Query 54 EAQGITTKSLTKVFVGGLAYSTSRYDLCELFA--------EVG--NIKSIVIPQVDR-SI 102
E+Q + +S ++VG L++ T++ L + F EV NIK I Q+ R +
Sbjct 95 ESQEESKRSPWGIWVGNLSFHTTKEILTDFFVRETSEMIKEVSEENIKPITTEQITRIHM 154
Query 103 PPDYEGAYSVRGIAFVQFEDAESAKKALQCNGKIVNGRSVVV 144
P E + +G A+V F ++ K ALQC+ K +NGR++++
Sbjct 155 PMSKEKRFQNKGFAYVDFATEDALKLALQCSEKALNGRNILI 196
Score = 39.3 bits (90), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 20/66 (30%), Positives = 33/66 (50%), Gaps = 8/66 (12%)
Query 66 VFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFEDAES 125
+FVG L + T+ DL E F +VG I+ + + +E +G FV F D ++
Sbjct 230 LFVGNLDFETTDADLKEHFGQVGQIRRVRLMT--------FEDTGKCKGFGFVDFPDIDT 281
Query 126 AKKALQ 131
KA++
Sbjct 282 CMKAME 287
> At4g39260
Length=169
Score = 51.6 bits (122), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 30/84 (35%), Positives = 48/84 (57%), Gaps = 9/84 (10%)
Query 65 KVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFEDAE 124
+ FVGGLA++T+ DL F++ G++ +D I D E S RG FV F+D +
Sbjct 7 RCFVGGLAWATNDEDLQRTFSQFGDV-------IDSKIINDRESGRS-RGFGFVTFKDEK 58
Query 125 SAKKAL-QCNGKIVNGRSVVVTPA 147
+ + A+ + NGK ++GR + V A
Sbjct 59 AMRDAIEEMNGKELDGRVITVNEA 82
> SPBC660.11
Length=348
Score = 51.6 bits (122), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 32/95 (33%), Positives = 50/95 (52%), Gaps = 15/95 (15%)
Query 65 KVFVGGLAYSTSRYDLCELFAEVGNIKSIVIP-----QVDRSIPPDYEGAYSVRGIAFVQ 119
+VFVG L+ ST + ++ LF VG ++ + IP + R +P GIAFV
Sbjct 45 RVFVGRLSTSTKKSEIRSLFETVGTVRKVTIPFRRVRRGTRLVP---------SGIAFVT 95
Query 120 FEDAESAKKALQC-NGKIVNGRSVVVTPANLTREE 153
F + E KA++ NGK ++ R +VV A +E+
Sbjct 96 FNNQEDVDKAIETLNGKTLDDREIVVQKARPVQEQ 130
> CE13934
Length=248
Score = 50.1 bits (118), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 37/111 (33%), Positives = 53/111 (47%), Gaps = 9/111 (8%)
Query 63 LTKVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFED 122
TK+FVGGL Y TS L E F + G+I+ V+ DR+ RG FV +D
Sbjct 34 FTKIFVGGLPYHTSDKTLHEYFEQFGDIEEAVV-ITDRN-------TQKSRGYGFVTMKD 85
Query 123 AESAKKALQCNGKIVNGRSVVVTPANLTREERA-AQEDLMAEGEMNFVAAT 172
SA++A + I++GR V A L + R Q +A G++ T
Sbjct 86 RASAERACKDPNPIIDGRKANVNLAYLGAKPRTNVQLAALAAGQVQLPLTT 136
> SPAC140.02
Length=500
Score = 49.3 bits (116), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 26/75 (34%), Positives = 41/75 (54%), Gaps = 8/75 (10%)
Query 66 VFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFEDAES 125
VFVG L+++ + DL F G+I+SI +P +S ++G +V F D +S
Sbjct 368 VFVGNLSFNATEDDLSTAFGGCGDIQSIRLPTDPQS--------GRLKGFGYVTFSDIDS 419
Query 126 AKKALQCNGKIVNGR 140
AKK ++ NG + GR
Sbjct 420 AKKCVEMNGHFIAGR 434
> At2g21660
Length=176
Score = 49.3 bits (116), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 31/84 (36%), Positives = 47/84 (55%), Gaps = 9/84 (10%)
Query 65 KVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFEDAE 124
+ FVGGLA++T L FA+ G++ +D I D E S RG FV F+D +
Sbjct 9 RCFVGGLAWATDDRALETAFAQYGDV-------IDSKIINDRETGRS-RGFGFVTFKDEK 60
Query 125 SAKKALQ-CNGKIVNGRSVVVTPA 147
+ K A++ NG+ ++GRS+ V A
Sbjct 61 AMKDAIEGMNGQDLDGRSITVNEA 84
> At5g06210
Length=146
Score = 48.9 bits (115), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 31/85 (36%), Positives = 50/85 (58%), Gaps = 15/85 (17%)
Query 64 TKVFVGGLAYSTSRYDLCELFAEVGNI--KSIVIPQV-DRSIPPDYEGAYSVRGIAFVQF 120
+K+F+GGL++ T+ L E F++ G + IV+ +V DRS +G FV F
Sbjct 34 SKLFIGGLSFCTTEQGLSEAFSKCGQVVEAQIVMDRVSDRS-----------KGFGFVTF 82
Query 121 EDAESAKKAL-QCNGKIVNGRSVVV 144
A+ A+KAL + NG+ +NGR++ V
Sbjct 83 ASADEAQKALMEFNGQQLNGRTIFV 107
> ECU03g0730
Length=293
Score = 48.5 bits (114), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 29/86 (33%), Positives = 46/86 (53%), Gaps = 9/86 (10%)
Query 62 SLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFE 121
SLT VF+ G++Y + YDL ++G + + IP + D++ +G +V+F
Sbjct 86 SLT-VFIKGISYDLTEYDLKSEMEKIGKVARVGIPMTN-----DHK---RNKGFGYVEFC 136
Query 122 DAESAKKALQCNGKIVNGRSVVVTPA 147
E KKAL+ +G + GR VVV A
Sbjct 137 KEEDVKKALKLDGTVFLGREVVVNMA 162
> 7300420
Length=399
Score = 48.1 bits (113), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 31/98 (31%), Positives = 53/98 (54%), Gaps = 10/98 (10%)
Query 48 LAEQVNEAQGITTKSLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYE 107
+A++ E Q I KS+ VFVG + Y + L E+F+EVG + S+ + DR
Sbjct 1 MADKAQE-QSIMDKSMRSVFVGNIPYEATEEKLKEIFSEVGPVLSLKL-VFDRE------ 52
Query 108 GAYSVRGIAFVQFEDAESAKKALQ-CNGKIVNGRSVVV 144
+ +G F +++D E+A A++ NG + GR++ V
Sbjct 53 -SGKPKGFGFCEYKDQETALSAMRNLNGYEIGGRTLRV 89
> At3g18610
Length=636
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 31/97 (31%), Positives = 51/97 (52%), Gaps = 13/97 (13%)
Query 52 VNEAQGITTKSLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYS 111
N+ QG +K+L F G L+Y +R D+ F E G + + + D +G++
Sbjct 376 TNQTQG-GSKTL---FAGNLSYQIARSDIENFFKEAGEVVDVRLSSFD-------DGSF- 423
Query 112 VRGIAFVQFEDAESAKKALQCNGKIVNGRSVVVTPAN 148
+G ++F E A+KAL+ NGK++ GR V + AN
Sbjct 424 -KGYGHIEFASPEEAQKALEMNGKLLLGRDVRLDLAN 459
> Hs20480107
Length=249
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 33/95 (34%), Positives = 50/95 (52%), Gaps = 9/95 (9%)
Query 62 SLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFE 121
++ K+FVGG+ T + L + F + G I+ I I DR G+ RG AFV F+
Sbjct 32 TVKKIFVGGIKEDTEEHHLRDYFEQYGKIEVIEI-MSDR-------GSGKKRGFAFVTFD 83
Query 122 DAESAKKALQCNGKIVNGRSVVVTPANLTREERAA 156
D +S K + VNG + VT A L+++E A+
Sbjct 84 DHDSVDKIVIQKYHTVNGHNCEVTKA-LSKQEMAS 117
> At3g54770
Length=261
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 34/95 (35%), Positives = 49/95 (51%), Gaps = 16/95 (16%)
Query 49 AEQVNEAQGITTKSLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQ---VDRSIPPD 105
+ VN G T LTKVFVGGLA+ T + + + F + G+I VI RS
Sbjct 4 SNNVNGCFGDT--KLTKVFVGGLAWDTHKEAMYDHFIKYGDILEAVIISDKLTRRS---- 57
Query 106 YEGAYSVRGIAFVQFEDAESAKKALQCNGKIVNGR 140
+G FV F+DA++A +A + + I+NGR
Sbjct 58 -------KGYGFVTFKDAKAATRACEDSTPIINGR 85
> Hs6857826
Length=305
Score = 47.4 bits (111), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 27/83 (32%), Positives = 49/83 (59%), Gaps = 8/83 (9%)
Query 64 TKVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFEDA 123
TKV +G L + ++ + E+F+ G IK I +P V+R P + +G A+V+FE+
Sbjct 161 TKVHIGRLTRNVTKDHIMEIFSTYGKIKMIDMP-VERMHP------HLSKGYAYVEFENP 213
Query 124 ESAKKALQ-CNGKIVNGRSVVVT 145
+ A+KAL+ +G ++G+ + T
Sbjct 214 DEAEKALKHMDGGQIDGQEITAT 236
> At2g46780
Length=304
Score = 47.4 bits (111), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 29/83 (34%), Positives = 46/83 (55%), Gaps = 10/83 (12%)
Query 59 TTKSLTKVFVGGLAYSTSRYDLCELFAEVGNI-KSIVIPQVDRSIPPDYEGAYSVRGIAF 117
T LTK+FVGGLA+ T R + F + G I +++VI D++ +G F
Sbjct 17 TDTKLTKIFVGGLAWETQRDTMRRYFEQFGEIVEAVVI--TDKNTG-------RSKGYGF 67
Query 118 VQFEDAESAKKALQCNGKIVNGR 140
V F++AE+A +A Q +++GR
Sbjct 68 VTFKEAEAAMRACQNMNPVIDGR 90
> 7297190
Length=421
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 32/116 (27%), Positives = 53/116 (45%), Gaps = 12/116 (10%)
Query 65 KVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFEDAE 124
K+FVGGL++ T++ +L F G+I V+ + + S RG FV F D
Sbjct 8 KLFVGGLSWETTQENLSRYFCRFGDIIDCVVMKNNESG--------RSRGFGFVTFADPT 59
Query 125 SAKKALQCNGKIVNGRSVVVTPAN---LTREERAAQEDLMAEGEMNFVAATD-KTF 176
+ LQ ++GR++ P N L + ++ + G + V TD +TF
Sbjct 60 NVNHVLQNGPHTLDGRTIDPKPCNPRTLQKPKKGGGYKVFLGGLPSNVTETDLRTF 115
Score = 32.3 bits (72), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 23/78 (29%), Positives = 34/78 (43%), Gaps = 13/78 (16%)
Query 65 KVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFEDAE 124
KVF+GGL + + DL F G + +VI D+ RG F+ FE+
Sbjct 97 KVFLGGLPSNVTETDLRTFFNRYGKVTEVVI-MYDQE-------KKKSRGFGFLSFEEES 148
Query 125 SA-----KKALQCNGKIV 137
S ++ + NGK V
Sbjct 149 SVEHVTNERYINLNGKQV 166
> Hs13650572
Length=321
Score = 46.6 bits (109), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 32/95 (33%), Positives = 49/95 (51%), Gaps = 9/95 (9%)
Query 62 SLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFE 121
++ K+FVGG+ T + L + F + G I+ I I DR G+ RG AFV F+
Sbjct 103 TVKKIFVGGIKEDTEEHHLRDYFEQYGKIEVIEI-MTDR-------GSGKKRGFAFVTFD 154
Query 122 DAESAKKALQCNGKIVNGRSVVVTPANLTREERAA 156
D +S K + VNG + V A L+++E A+
Sbjct 155 DHDSVDKTVIQKYHTVNGHNCEVRKA-LSKQEMAS 188
Score = 36.2 bits (82), Expect = 0.049, Method: Compositional matrix adjust.
Identities = 28/95 (29%), Positives = 42/95 (44%), Gaps = 17/95 (17%)
Query 63 LTKVFVGGLAYSTSRYDLCELFAEVGNIKSIVI---PQVDRSIPPDYEGAYSVRGIAFVQ 119
L K+F+GGL++ T+ L F + G + V+ P RS RG FV
Sbjct 13 LRKLFIGGLSFETTDESLRSHFEQWGTLTDCVVMRDPNTKRS-----------RGFGFVT 61
Query 120 FEDAESAKKALQCNGKIVNGRSVVVTPAN-LTREE 153
+ E A+ V+GR VV P ++RE+
Sbjct 62 YATVEEVDAAMNARPHKVDGR--VVEPKRAVSRED 94
> Hs22067735
Length=268
Score = 46.6 bits (109), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 32/95 (33%), Positives = 49/95 (51%), Gaps = 9/95 (9%)
Query 62 SLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFE 121
++ K+FVGG+ T + L + F + G I+ I I DR G+ RG AFV F+
Sbjct 103 TVKKIFVGGIKEDTEEHHLRDYFEQYGKIEVIEI-MTDR-------GSGKKRGFAFVTFD 154
Query 122 DAESAKKALQCNGKIVNGRSVVVTPANLTREERAA 156
D +S K + VNG + V A L+++E A+
Sbjct 155 DHDSVDKIVIQKYHTVNGHNCEVRKA-LSKQEMAS 188
Score = 37.4 bits (85), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 28/100 (28%), Positives = 41/100 (41%), Gaps = 16/100 (16%)
Query 63 LTKVFVGGLAYSTSRYDLCELFAEVGNIKSIVI---PQVDRSIPPDYEGAYSVRGIAFVQ 119
L K+F+GGL++ T+ L F + G + V+ P RS RG FV
Sbjct 13 LRKLFIGGLSFETTDESLRSHFEQWGTLTDCVVMRDPNTKRS-----------RGFGFVT 61
Query 120 FEDAESAKKALQCNGKIVNGRSVVVTPANLTREERAAQED 159
+ E A+ V+GR VV P E + + D
Sbjct 62 YATVEEVDAAMNARPHKVDGR--VVEPKRAVSREDSQRPD 99
> 7299789
Length=329
Score = 46.2 bits (108), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 30/88 (34%), Positives = 39/88 (44%), Gaps = 17/88 (19%)
Query 64 TKVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFEDA 123
++V+VGGL Y DL F G + I+I G FV+FED
Sbjct 4 SRVYVGGLPYGVRERDLERFFKGYGRTRDILIKN----------------GYGFVEFEDY 47
Query 124 ESAKKAL-QCNGKIVNGRSVVVTPANLT 150
A A+ + NGK + G VVV PA T
Sbjct 48 RDADDAVYELNGKELLGERVVVEPARGT 75
> Hs14043070
Length=372
Score = 46.2 bits (108), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 32/95 (33%), Positives = 49/95 (51%), Gaps = 9/95 (9%)
Query 62 SLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFE 121
++ K+FVGG+ T + L + F + G I+ I I DR G+ RG AFV F+
Sbjct 103 TVKKIFVGGIKEDTEEHHLRDYFEQYGKIEVIEI-MTDR-------GSGKKRGFAFVTFD 154
Query 122 DAESAKKALQCNGKIVNGRSVVVTPANLTREERAA 156
D +S K + VNG + V A L+++E A+
Sbjct 155 DHDSVDKIVIQKYHTVNGHNCEVRKA-LSKQEMAS 188
Score = 36.6 bits (83), Expect = 0.046, Method: Compositional matrix adjust.
Identities = 28/95 (29%), Positives = 42/95 (44%), Gaps = 17/95 (17%)
Query 63 LTKVFVGGLAYSTSRYDLCELFAEVGNIKSIVI---PQVDRSIPPDYEGAYSVRGIAFVQ 119
L K+F+GGL++ T+ L F + G + V+ P RS RG FV
Sbjct 13 LRKLFIGGLSFETTDESLRSHFEQWGTLTDCVVMRDPNTKRS-----------RGFGFVT 61
Query 120 FEDAESAKKALQCNGKIVNGRSVVVTPAN-LTREE 153
+ E A+ V+GR VV P ++RE+
Sbjct 62 YATVEEVDAAMNARPHKVDGR--VVEPKRAVSRED 94
> Hs4504445
Length=320
Score = 45.8 bits (107), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 32/95 (33%), Positives = 49/95 (51%), Gaps = 9/95 (9%)
Query 62 SLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFE 121
++ K+FVGG+ T + L + F + G I+ I I DR G+ RG AFV F+
Sbjct 103 TVKKIFVGGIKEDTEEHHLRDYFEQYGKIEVIEI-MTDR-------GSGKKRGFAFVTFD 154
Query 122 DAESAKKALQCNGKIVNGRSVVVTPANLTREERAA 156
D +S K + VNG + V A L+++E A+
Sbjct 155 DHDSVDKIVIQKYHTVNGHNCEVRKA-LSKQEMAS 188
Score = 36.2 bits (82), Expect = 0.050, Method: Compositional matrix adjust.
Identities = 28/95 (29%), Positives = 42/95 (44%), Gaps = 17/95 (17%)
Query 63 LTKVFVGGLAYSTSRYDLCELFAEVGNIKSIVI---PQVDRSIPPDYEGAYSVRGIAFVQ 119
L K+F+GGL++ T+ L F + G + V+ P RS RG FV
Sbjct 13 LRKLFIGGLSFETTDESLRSHFEQWGTLTDCVVMRDPNTKRS-----------RGFGFVT 61
Query 120 FEDAESAKKALQCNGKIVNGRSVVVTPAN-LTREE 153
+ E A+ V+GR VV P ++RE+
Sbjct 62 YATVEEVDAAMNARPHKVDGR--VVEPKRAVSRED 94
> Hs14110404
Length=331
Score = 45.8 bits (107), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 29/85 (34%), Positives = 42/85 (49%), Gaps = 16/85 (18%)
Query 65 KVFVGGLAYSTSRYDLCELFAEVGNIKSIVI---PQVDRSIPPDYEGAYSVRGIAFVQFE 121
K+FVGGL++ TS+ DL + F + G + I P RS RG F+ F+
Sbjct 69 KMFVGGLSWDTSKKDLKDYFTKFGEVVDCTIKMDPNTGRS-----------RGFGFILFK 117
Query 122 DAESAKKALQCNGKIVNGRSVVVTP 146
DA S +K L ++GR V+ P
Sbjct 118 DAASVEKVLDQKEHRLDGR--VIDP 140
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 36/137 (26%), Positives = 60/137 (43%), Gaps = 16/137 (11%)
Query 4 VQMQTNDSFSLGASSRHYLLEIDTTSVVDYLQRRGRELNRELSLLAEQVNEAQGITTKSL 63
++M N S G ++L D SV L ++ L+ + +A + +
Sbjct 99 IKMDPNTGRSRGFG---FILFKDAASVEKVLDQKEHRLDGRVI----DPKKAMAMKKDPV 151
Query 64 TKVFVGGL-AYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFED 122
K+FVGGL S + + E F E G I++I +P +D + RG F+ F++
Sbjct 152 KKIFVGGLNPESPTEEKIREYFGEFGEIEAIELP-MDPKL-------NKRRGFVFITFKE 203
Query 123 AESAKKALQCNGKIVNG 139
E KK L+ V+G
Sbjct 204 EEPVKKVLEKKFHTVSG 220
> At1g20880
Length=277
Score = 45.8 bits (107), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 36/113 (31%), Positives = 54/113 (47%), Gaps = 12/113 (10%)
Query 52 VNEAQGITTKSLTKVFVGGLAYSTSRYDLCELFAEVGNI-KSIVIPQVDRSIPPDYEGAY 110
+N G TT TKVFVGGLA+ T L F + G+I +++VI D++
Sbjct 14 LNSPFGDTT--FTKVFVGGLAWETQSETLRRHFDQYGDILEAVVI--TDKN-------TG 62
Query 111 SVRGIAFVQFEDAESAKKALQCNGKIVNGRSVVVTPANLTREERAAQEDLMAE 163
+G FV F D E+A++A I++GR A+L R Q ++
Sbjct 63 RSKGYGFVTFRDPEAARRACVDPTPIIDGRRANCNLASLGRSRPPMQYAVIPH 115
> SPBC18H10.04c
Length=388
Score = 45.4 bits (106), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 42/155 (27%), Positives = 69/155 (44%), Gaps = 22/155 (14%)
Query 40 ELNRELSLLAEQVNEAQGITTKSLTKVFVGGLAYSTSRYDLCELFAE-VGNIKSIVIPQV 98
E RE + + +A I ++ VG L++ + DL + F E V +I+ ++ P
Sbjct 70 ESRREGGMGSGYQRDAIPIPSEPPFTAHVGNLSFDLTENDLGDFFGEGVTSIRLVIDPLT 129
Query 99 DRSIPPDYEGAYSVRGIAFVQFEDAESAKKALQCNGKIVNGRSVVVTPA----NLTREER 154
+RS RG +V+FE A++ AL +G+ + GR V +T A + REER
Sbjct 130 ERS-----------RGFGYVEFETADTLSAALALSGEDLMGRPVRITVAEPRRSFAREER 178
Query 155 AAQEDLMAEGEMNFVAATDKTFDHPTHGSLRTGHG 189
+ D + G + + F G RT G
Sbjct 179 STG-DWVRRGPLPPAEPAESPF-----GKRRTNSG 207
> Hs4758542
Length=284
Score = 45.4 bits (106), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 27/81 (33%), Positives = 40/81 (49%), Gaps = 14/81 (17%)
Query 65 KVFVGGLAYSTSRYDLCELFAEVGNIKSIVI---PQVDRSIPPDYEGAYSVRGIAFVQFE 121
K+FVGGL++ TS+ DL + F + G + I P RS RG F+ F+
Sbjct 69 KMFVGGLSWDTSKKDLKDYFTKFGEVVDCTIKMDPNTGRS-----------RGFGFILFK 117
Query 122 DAESAKKALQCNGKIVNGRSV 142
DA S +K L ++GR +
Sbjct 118 DAASVEKVLDQKEHRLDGRVI 138
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 36/137 (26%), Positives = 60/137 (43%), Gaps = 16/137 (11%)
Query 4 VQMQTNDSFSLGASSRHYLLEIDTTSVVDYLQRRGRELNRELSLLAEQVNEAQGITTKSL 63
++M N S G ++L D SV L ++ L+ + +A + +
Sbjct 99 IKMDPNTGRSRGFG---FILFKDAASVEKVLDQKEHRLDGRVI----DPKKAMAMKKDPV 151
Query 64 TKVFVGGL-AYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFED 122
K+FVGGL S + + E F E G I++I +P +D + RG F+ F++
Sbjct 152 KKIFVGGLNPESPTEEKIREYFGEFGEIEAIELP-MDPKL-------NKRRGFVFITFKE 203
Query 123 AESAKKALQCNGKIVNG 139
E KK L+ V+G
Sbjct 204 EEPVKKVLEKKFHTVSG 220
> YGR159c
Length=414
Score = 45.4 bits (106), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 26/78 (33%), Positives = 42/78 (53%), Gaps = 9/78 (11%)
Query 66 VFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFEDAES 125
+F+G L+++ R + ELFA+ G + S+ IP + P +G +VQF + E
Sbjct 269 LFLGNLSFNADRDAIFELFAKHGEVVSVRIPTHPETEQP--------KGFGYVQFSNMED 320
Query 126 AKKALQC-NGKIVNGRSV 142
AKKAL G+ ++ R V
Sbjct 321 AKKALDALQGEYIDNRPV 338
Score = 34.3 bits (77), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 25/81 (30%), Positives = 39/81 (48%), Gaps = 15/81 (18%)
Query 66 VFVGGLAYSTSRYDLCELFAEVGNI---KSIVIPQVDRSIPPDYEGAYSVRGIAFVQFED 122
+FVG L++S L + F +G + + I DRS RG +V FE+
Sbjct 170 IFVGRLSWSIDDEWLKKEFEHIGGVIGARVIYERGTDRS-----------RGYGYVDFEN 218
Query 123 AESAKKALQ-CNGKIVNGRSV 142
A+KA+Q GK ++GR +
Sbjct 219 KSYAEKAIQEMQGKEIDGRPI 239
> At2g23350
Length=662
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 34/97 (35%), Positives = 48/97 (49%), Gaps = 11/97 (11%)
Query 66 VFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFEDAES 125
+FV L S L E F+ G I S + D+ G RG FVQF+ +S
Sbjct 136 LFVKNLDKSVDNKTLHEAFSGCGTIVSC-------KVATDHMG--QSRGYGFVQFDTEDS 186
Query 126 AKKALQ-CNGKIVNGRSVVVTPANLTREERAAQEDLM 161
AK A++ NGK++N + + V P L +EER + D M
Sbjct 187 AKNAIEKLNGKVLNDKQIFVGPF-LRKEERESAADKM 222
Score = 33.5 bits (75), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 26/91 (28%), Positives = 42/91 (46%), Gaps = 10/91 (10%)
Query 63 LTKVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFED 122
T V+V L+ +T+ +L F + G+I S V+ + +G R FV FE+
Sbjct 224 FTNVYVKNLSEATTDDELKTTFGQYGSISSAVVMR---------DGDGKSRCFGFVNFEN 274
Query 123 AESAKKALQC-NGKIVNGRSVVVTPANLTRE 152
E A +A++ NGK + + V A E
Sbjct 275 PEDAARAVEALNGKKFDDKEWYVGKAQKKSE 305
Score = 29.6 bits (65), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 21/85 (24%), Positives = 42/85 (49%), Gaps = 9/85 (10%)
Query 62 SLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFE 121
+L ++VG L ++ + L + F EV + S+ +V R + G +V +
Sbjct 44 ALCSLYVGDLDFNVTDSQLYDYFTEVCQVVSV---RVCRDA-----ATNTSLGYGYVNYS 95
Query 122 DAESAKKALQ-CNGKIVNGRSVVVT 145
+ + A+KA+Q N +NG+ + +T
Sbjct 96 NTDDAEKAMQKLNYSYLNGKMIRIT 120
> CE01888
Length=237
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 35/132 (26%), Positives = 66/132 (50%), Gaps = 16/132 (12%)
Query 17 SSRHYLLEIDTTSVVDYLQRRGRELNREL---SLLAEQVNEAQGITTKSLTKVFVGGLAY 73
S+ H+ +EI+ S + LQ+ ++ + L + + E + I KS VF+G + +
Sbjct 56 SNSHFEMEIEAESAI--LQQIQNKMAKHLESAAYVPPTEEEQKAIDAKS---VFIGNVDF 110
Query 74 STSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFEDAESAKKALQCN 133
+++ ++ E F G I V +IP D + + A+++F+D+ S + AL N
Sbjct 111 NSTIEEIEEHFKGCGQI-------VKTTIPKD-KFTKKQKNFAYIEFDDSSSIENALVMN 162
Query 134 GKIVNGRSVVVT 145
G + R +VVT
Sbjct 163 GSLFRSRPIVVT 174
> CE01891
Length=196
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 29/90 (32%), Positives = 45/90 (50%), Gaps = 9/90 (10%)
Query 63 LTKVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFED 122
LT + + L+Y T+ DL F G+I + IP+ S +G FV+F +
Sbjct 18 LTSLKIDNLSYQTTPNDLRRTFERYGDIGDVHIPRDKYS--------RQSKGFGFVRFYE 69
Query 123 AESAKKAL-QCNGKIVNGRSVVVTPANLTR 151
A+ AL + +GK+V+GR + VT A R
Sbjct 70 RRDAEHALDRTDGKLVDGRELRVTLAKYDR 99
> CE27910
Length=126
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 29/90 (32%), Positives = 45/90 (50%), Gaps = 9/90 (10%)
Query 63 LTKVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFED 122
LT + + L+Y T+ DL F G+I + IP+ S +G FV+F +
Sbjct 18 LTSLKIDNLSYQTTPNDLRRTFERYGDIGDVHIPRDKYS--------RQSKGFGFVRFYE 69
Query 123 AESAKKAL-QCNGKIVNGRSVVVTPANLTR 151
A+ AL + +GK+V+GR + VT A R
Sbjct 70 RRDAEHALDRTDGKLVDGRELRVTLAKYDR 99
> Hs14733321
Length=305
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 27/83 (32%), Positives = 48/83 (57%), Gaps = 8/83 (9%)
Query 64 TKVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFEDA 123
TKV +G L + ++ + E+F+ G IK I +P V+R P + +G A V+FE+
Sbjct 161 TKVHIGRLTRNVTKDHIMEIFSTYGKIKMIDMP-VERMHP------HLSKGYACVEFENP 213
Query 124 ESAKKALQ-CNGKIVNGRSVVVT 145
+ A+KAL+ +G ++G+ + T
Sbjct 214 DEAEKALKHMDGGQIDGQEITAT 236
> At1g18630
Length=155
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 34/94 (36%), Positives = 47/94 (50%), Gaps = 9/94 (9%)
Query 56 QGITTKSLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGI 115
QG + + +K+FVGGL+ ST L E F G I VD + D E S RG
Sbjct 28 QGNLSLTPSKIFVGGLSPSTDVELLKEAFGSFGKI-------VDAVVVLDRESGLS-RGF 79
Query 116 AFVQFEDAESAKKALQC-NGKIVNGRSVVVTPAN 148
FV ++ E A A+Q K ++GR + V PA+
Sbjct 80 GFVTYDSIEVANNAMQAMQNKELDGRIIGVHPAD 113
> At1g33470
Length=117
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 29/80 (36%), Positives = 45/80 (56%), Gaps = 10/80 (12%)
Query 62 SLTKVFVGGLAYSTSRYDLCELFAEVGNI-KSIVIPQVDRSIPPDYEGAYSVRGIAFVQF 120
+ TKVFVGGLA+ T + L F + G+I +++VI D+S + +G FV F
Sbjct 5 TFTKVFVGGLAWETHKVSLRNYFEQFGDIVEAVVI--TDKS-------SGRSKGYGFVTF 55
Query 121 EDAESAKKALQCNGKIVNGR 140
D E+A+KA +++GR
Sbjct 56 CDPEAAQKACVDPAPVIDGR 75
> YFR023w
Length=611
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 32/104 (30%), Positives = 55/104 (52%), Gaps = 14/104 (13%)
Query 54 EAQGITTKSLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVR 113
E++ I + S +F+ L T+R D+ F+EVG IKSI + A V+
Sbjct 293 ESKNIYSSSQNSIFIKNLPTITTRDDILNFFSEVGPIKSIYL-----------SNATKVK 341
Query 114 GI-AFVQFEDAESAKKALQ-CNGKIVNGRSVVVTPANLTREERA 155
+ AFV ++++ ++KA++ N G+ ++VT A +EERA
Sbjct 342 YLWAFVTYKNSSDSEKAIKRYNNFYFRGKKLLVTRAQ-DKEERA 384
> At5g53680
Length=169
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 26/82 (31%), Positives = 40/82 (48%), Gaps = 14/82 (17%)
Query 62 SLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIVI---PQVDRSIPPDYEGAYSVRGIAFV 118
+ TK++VGGL ++T + L F G I + + + DRS +G FV
Sbjct 11 TFTKIYVGGLPWTTRKEGLINFFKRFGEIIHVNVVCDRETDRS-----------QGYGFV 59
Query 119 QFEDAESAKKALQCNGKIVNGR 140
F+DAESA +A + + GR
Sbjct 60 TFKDAESATRACKDPNPTIEGR 81
> Hs22049560
Length=222
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 32/95 (33%), Positives = 48/95 (50%), Gaps = 9/95 (9%)
Query 62 SLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFE 121
++ K+FVGG+ T + L + F + G I+ I I DR G+ RG AFV F+
Sbjct 103 TVKKIFVGGIKEDTKEHHLRDYFEQYGKIEVIEI-MTDR-------GSGKKRGFAFVTFD 154
Query 122 DAESAKKALQCNGKIVNGRSVVVTPANLTREERAA 156
D +S K + VNG V A L+++E A+
Sbjct 155 DHDSVDKIVIQKYHTVNGHICEVRKA-LSKQEMAS 188
Score = 36.2 bits (82), Expect = 0.062, Method: Compositional matrix adjust.
Identities = 26/93 (27%), Positives = 45/93 (48%), Gaps = 9/93 (9%)
Query 61 KSLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQF 120
K L K+F+GGL++ T+ L F + G + V+ + P+ + + RG FV +
Sbjct 11 KQLRKLFIGGLSFETTNESLRSHFEQWGTLMDCVVMR-----DPNTKCS---RGFGFVTY 62
Query 121 EDAESAKKALQCNGKIVNGRSVVVTPANLTREE 153
E A+ V+GR VV + ++RE+
Sbjct 63 ATVEEVDAAMNARPHKVDGR-VVESKRAVSRED 94
> Hs14192947
Length=216
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 32/96 (33%), Positives = 47/96 (48%), Gaps = 8/96 (8%)
Query 62 SLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFE 121
+ TK+FVGGL Y T+ L + F G+I+ V+ DR G RG FV
Sbjct 9 TFTKIFVGGLPYHTTDASLRKYFEGFGDIEEAVV-ITDRQ-----TG--KSRGYGFVTMA 60
Query 122 DAESAKKALQCNGKIVNGRSVVVTPANLTREERAAQ 157
D +A++A + I++GR V A L + R+ Q
Sbjct 61 DRAAAERACKDPNPIIDGRKANVNLAYLGAKPRSLQ 96
> Hs14731484
Length=320
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 31/92 (33%), Positives = 47/92 (51%), Gaps = 9/92 (9%)
Query 62 SLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFE 121
++ K+FVGG+ T + L + F + G I+ I I DR G+ RG AFV F+
Sbjct 103 TVKKIFVGGIKEDTEEHHLRDYFEQYGKIEVIEI-MTDR-------GSGKKRGFAFVTFD 154
Query 122 DAESAKKALQCNGKIVNGRSVVVTPANLTREE 153
D +S K + VNG + V A L+++E
Sbjct 155 DHDSVDKIVIQKYHTVNGHNCEVRKA-LSKQE 185
Score = 29.6 bits (65), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 25/95 (26%), Positives = 42/95 (44%), Gaps = 17/95 (17%)
Query 63 LTKVFVGGLAYSTSRYDLCELFAEVGNIKSIVI---PQVDRSIPPDYEGAYSVRGIAFVQ 119
L K+F+GGL++ T+ L F + G + V+ P S G FV
Sbjct 13 LRKLFIGGLSFETTDESLRSHFEQRGTLTDCVVMRDPNTKCST-----------GFGFVT 61
Query 120 FEDAESAKKALQCNGKIVNGRSVVVTPAN-LTREE 153
+ + + A+ + V+GR VV P ++RE+
Sbjct 62 YATVKEVEAAMNARPQKVDGR--VVEPKRAVSRED 94
> YPR112c
Length=887
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 32/105 (30%), Positives = 53/105 (50%), Gaps = 13/105 (12%)
Query 44 ELSLLAEQVNEAQGITTKSLTK---VFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDR 100
+L L Q +++ TKS K + V L + +R D+ ELF G +KS+ +P+
Sbjct 740 QLKLSHRQASQSGNTKTKSNKKSGKIIVKNLPFEATRKDVFELFNSFGQLKSVRVPK--- 796
Query 101 SIPPDYEGAYSVRGIAFVQFEDAESAKKAL-QCNGKIVNGRSVVV 144
+ S RG AFV+F + A+ A+ Q +G + GR +V+
Sbjct 797 ------KFDKSARGFAFVEFLLPKEAENAMDQLHGVHLLGRRLVM 835
> CE29008
Length=305
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 29/90 (32%), Positives = 44/90 (48%), Gaps = 9/90 (10%)
Query 65 KVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFEDAE 124
K+FVGG++ + DL F + G + + + DR+ RG AFV+F E
Sbjct 30 KIFVGGISPEVNNEDLSSHFTQYGEVAQAQV-KYDRTNG-------RSRGFAFVEFTTGE 81
Query 125 SAKKALQCNGKIVNGRSVVVTPANLTREER 154
K AL + + G+SV V PA +RE +
Sbjct 82 GCKLALAAREQTIKGKSVEVKPAK-SRENK 110
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 29/87 (33%), Positives = 41/87 (47%), Gaps = 9/87 (10%)
Query 43 RELSLLAEQVNEAQGITTKSLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSI 102
RE ++ + V E + ++ KVFVGGL S DL F + G + I P D+
Sbjct 90 REQTIKGKSV-EVKPAKSRENKKVFVGGLPSDYSEQDLRSHFEQFGKVDDIEWP-FDKQT 147
Query 103 PPDYEGAYSVRGIAFVQFEDAESAKKA 129
+ R AF+ FE+ ESA KA
Sbjct 148 K-------ARRNFAFIVFEEEESADKA 167
> At5g47620
Length=404
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 26/80 (32%), Positives = 40/80 (50%), Gaps = 8/80 (10%)
Query 65 KVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFEDAE 124
K+FVGGLA S + + + FA+ G I +V+ R+ P RG F+ ++ E
Sbjct 107 KIFVGGLASSVTEAEFKKYFAQFGMITDVVVMYDHRTQRP--------RGFGFISYDSEE 158
Query 125 SAKKALQCNGKIVNGRSVVV 144
+ K LQ +NG+ V V
Sbjct 159 AVDKVLQKTFHELNGKMVEV 178
Score = 33.9 bits (76), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 25/83 (30%), Positives = 39/83 (46%), Gaps = 12/83 (14%)
Query 65 KVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFEDAE 124
K+F+GG+++ TS L + F G + VI + DR+ RG FV F D
Sbjct 7 KLFIGGISWETSEDRLRDYFHSFGEVLEAVIMK-DRATG-------RARGFGFVVFADPN 58
Query 125 SAKKALQ----CNGKIVNGRSVV 143
A++ + +GKIV + V
Sbjct 59 VAERVVLLKHIIDGKIVEAKKAV 81
> Hs4885423
Length=420
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 26/81 (32%), Positives = 39/81 (48%), Gaps = 14/81 (17%)
Query 65 KVFVGGLAYSTSRYDLCELFAEVGNIKSIVI---PQVDRSIPPDYEGAYSVRGIAFVQFE 121
K+F+GGL++ TS+ DL E + G + I P RS RG FV F+
Sbjct 149 KMFIGGLSWDTSKKDLTEYLSRFGEVVDCTIKTDPVTGRS-----------RGFGFVLFK 197
Query 122 DAESAKKALQCNGKIVNGRSV 142
DA S K L+ ++G+ +
Sbjct 198 DAASVDKVLELKEHKLDGKLI 218
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 31/111 (27%), Positives = 53/111 (47%), Gaps = 11/111 (9%)
Query 21 YLLEIDTTSVVDYLQRRGRELNRELSLLAEQVNEAQGITTKSLTKVFVGGLAYSTSRYDL 80
++L D SV L+ + +L+ +L + ++ +G + KVFVGGL+ TS +
Sbjct 193 FVLFKDAASVDKVLELKEHKLDGKL-IDPKRAKALKG--KEPPKKVFVGGLSPDTSEEQI 249
Query 81 CELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFEDAESAKKALQ 131
E F G I++I +P ++ RG F+ + D E KK L+
Sbjct 250 KEYFGAFGEIENIELPMDTKT--------NERRGFCFITYTDEEPVKKLLE 292
> Hs22042102
Length=378
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 28/92 (30%), Positives = 49/92 (53%), Gaps = 9/92 (9%)
Query 62 SLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFE 121
++ K+FVGG+ T Y+L + F + G I++I + + DR + RG AFV F+
Sbjct 124 TVKKIFVGGIKEDTEEYNLRDYFEKYGKIETIEVME-DRQ-------SGKKRGFAFVTFD 175
Query 122 DAESAKKALQCNGKIVNGRSVVVTPANLTREE 153
D ++ K + +NG + V A L+++E
Sbjct 176 DHDTVDKIVVQKYHTINGHNCEVKKA-LSKQE 206
Score = 38.9 bits (89), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 30/95 (31%), Positives = 44/95 (46%), Gaps = 17/95 (17%)
Query 63 LTKVFVGGLAYSTSRYDLCELFAEVGNIKSIVI---PQVDRSIPPDYEGAYSVRGIAFVQ 119
L K+F+GGL++ T+ L E F + G + V+ PQ RS RG FV
Sbjct 34 LRKLFIGGLSFETTDDSLREHFEKWGTLTDCVVMRDPQTKRS-----------RGFGFVT 82
Query 120 FEDAESAKKALQCNGKIVNGRSVVVTPAN-LTREE 153
+ E A+ V+GR VV P ++RE+
Sbjct 83 YSCVEEVDAAMCARPHKVDGR--VVEPKRAVSRED 115
> 7299199
Length=374
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 28/91 (30%), Positives = 54/91 (59%), Gaps = 12/91 (13%)
Query 65 KVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQF---E 121
++ VG L + ++ + E+F+ G++K++ P VDR P++ RG+AFV++ E
Sbjct 220 RIHVGRLTRNVTKDHVFEIFSSFGDVKNVEFP-VDR-FHPNF-----GRGVAFVEYATPE 272
Query 122 DAESAKKALQCNGKIVNGRSVVVTPANLTRE 152
D ESA K + +G ++G+ + V+P L ++
Sbjct 273 DCESAMKHM--DGGQIDGQEITVSPVVLVKQ 301
> At1g76460
Length=279
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 36/104 (34%), Positives = 50/104 (48%), Gaps = 16/104 (15%)
Query 52 VNEAQGITTKSLTKVFVGGLAYSTSRYDLCELFAEVGNI-KSIVI--PQVDRSIPPDYEG 108
+N G TT TKVFVGGLA+ T L + F + G I +++VI RS
Sbjct 14 LNSPFGDTT--FTKVFVGGLAWETQSETLRQHFEQYGEILEAVVIADKNTGRS------- 64
Query 109 AYSVRGIAFVQFEDAESAKKALQCNGKIVNGRSVVVTPANLTRE 152
+G FV F D E+A++A I++GR A+L R
Sbjct 65 ----KGYGFVTFRDPEAARRACADPTPIIDGRRANCNLASLGRP 104
> Hs7705365
Length=960
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 30/105 (28%), Positives = 53/105 (50%), Gaps = 12/105 (11%)
Query 65 KVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFEDAE 124
++FV L Y+++ DL +LF++ G + + P + P +G AF+ F E
Sbjct 403 RLFVRNLPYTSTEEDLEKLFSKYGPLSELHYPIDSLTKKP--------KGFAFITFMFPE 454
Query 125 SAKKAL-QCNGKIVNGRSVVVTPANLTREERAAQEDLMAEGEMNF 168
A KA + +G++ GR + V P+ + +E A ED A G ++
Sbjct 455 HAVKAYSEVDGQVFQGRMLHVLPSTIKKE---ASEDASALGSSSY 496
Score = 42.7 bits (99), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 26/80 (32%), Positives = 41/80 (51%), Gaps = 6/80 (7%)
Query 66 VFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFEDAES 125
+F+ L + T+ L E+F++VG +KS I S + G G FV++ E
Sbjct 732 LFIKNLNFDTTEEKLKEVFSKVGTVKSCSI-----SKKKNKAGVLLSMGFGFVEYRKPEQ 786
Query 126 AKKAL-QCNGKIVNGRSVVV 144
A+KAL Q G +V+G + V
Sbjct 787 AQKALKQLQGHVVDGHKLEV 806
Score = 37.4 bits (85), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 22/90 (24%), Positives = 44/90 (48%), Gaps = 9/90 (10%)
Query 61 KSLTKVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQF 120
++ +K+ V + + ++ ELF+ G +K++ +P+ G + RG FV F
Sbjct 829 QTTSKILVRNIPFQAHSREIRELFSTFGELKTVRLPK-------KMTGTGTHRGFGFVDF 881
Query 121 EDAESAKKALQ--CNGKIVNGRSVVVTPAN 148
+ AK+A C+ + GR +V+ A+
Sbjct 882 LTKQDAKRAFNALCHSTHLYGRRLVLEWAD 911
Score = 31.6 bits (70), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 13/31 (41%), Positives = 19/31 (61%), Gaps = 0/31 (0%)
Query 114 GIAFVQFEDAESAKKALQCNGKIVNGRSVVV 144
G FV F + E K+AL+CN + + GR + V
Sbjct 334 GYIFVDFSNEEEVKQALKCNREYMGGRYIEV 364
> Hs7662250
Length=960
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 30/105 (28%), Positives = 53/105 (50%), Gaps = 12/105 (11%)
Query 65 KVFVGGLAYSTSRYDLCELFAEVGNIKSIVIPQVDRSIPPDYEGAYSVRGIAFVQFEDAE 124
++FV L Y+++ DL +LF++ G + + P + P +G AF+ F E
Sbjct 403 RLFVRNLPYTSTEEDLEKLFSKYGPLSELHYPIDSLTKKP--------KGFAFITFMFPE 454
Query 125 SAKKAL-QCNGKIVNGRSVVVTPANLTREERAAQEDLMAEGEMNF 168
A KA + +G++ GR + V P+ + +E A ED A G ++
Sbjct 455 HAVKAYSEVDGQVFQGRMLHVLPSTIKKE---ASEDASALGSSSY 496
Lambda K H
0.314 0.130 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4312910206
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40