bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
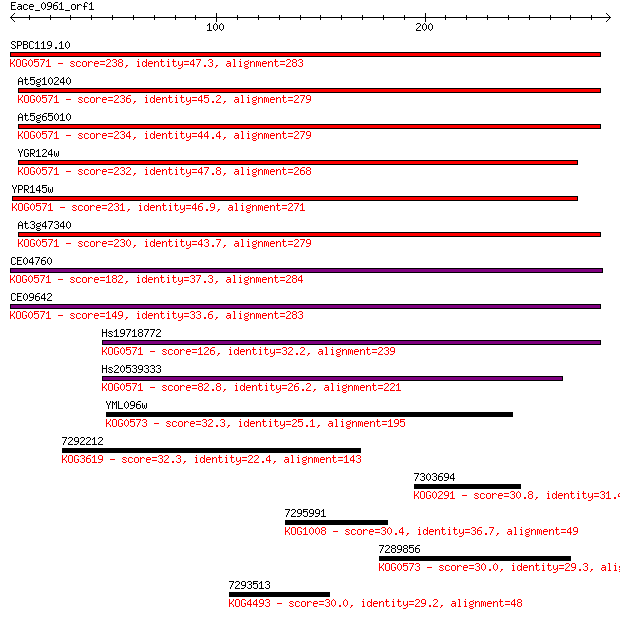
Query= Eace_0961_orf1
Length=288
Score E
Sequences producing significant alignments: (Bits) Value
SPBC119.10 238 7e-63
At5g10240 236 6e-62
At5g65010 234 1e-61
YGR124w 232 8e-61
YPR145w 231 2e-60
At3g47340 230 3e-60
CE04760 182 8e-46
CE09642 149 5e-36
Hs19718772 126 5e-29
Hs20539333 82.8 8e-16
YML096w 32.3 1.2
7292212 32.3 1.3
7303694 30.8 3.7
7295991 30.4 4.7
7289856 30.0 6.1
7293513 30.0 6.4
> SPBC119.10
Length=557
Score = 238 bits (608), Expect = 7e-63, Method: Compositional matrix adjust.
Identities = 134/296 (45%), Positives = 181/296 (61%), Gaps = 18/296 (6%)
Query 1 RVREFPPGHSFYFKSSKASGIMERFYKPQWMQPEAPIPSAACDLSRLREELEKAVSKRMM 60
++ FPPGH Y+ S + R++KP W E IPS D LRE LE +V KR+M
Sbjct 173 KIIAFPPGH--YYDSETKQTV--RYFKPSWWD-ENKIPSNPVDYKLLRETLEASVRKRLM 227
Query 61 CDVPFGVLLYGGIDSAIIASIVCRLYNERFNK-EQNEHFWT----PKIHSFAIGLKGSRD 115
+VP+GVLL GG+DS++IASI R + N Q+E T PK+HSFAIGL GS D
Sbjct 228 AEVPYGVLLSGGLDSSLIASIAARETEKLANSTSQSEEARTITAWPKLHSFAIGLPGSPD 287
Query 116 LECSRMVAKHVGCTHHEFTFEAQEAIDALSDLIYMIESYDVLTVRAALLNYFLYRLIKSI 175
L +R VA + HHE TF E +DAL D+IY +E+YDV T+RA+ Y L R IK+
Sbjct 288 LLAARKVADFLHTFHHEHTFTIDEGLDALRDVIYHLETYDVTTIRASTPMYLLSRKIKAQ 347
Query 176 GVKMVITGEGAADLFGCAGDMQEEQDPEALHKAIVKRMSDMHFTDSLRANKASMCWGVEA 235
GVKMV++GEG+ ++FG EA H V+R+ ++H +D LRANK++M WG+EA
Sbjct 348 GVKMVLSGEGSDEIFGGYLYFGNAPSREAFHSECVRRVKNLHLSDCLRANKSTMAWGLEA 407
Query 236 RVPYLDVSFVDYAMSIDPKEKRCSGGRLPKQLLRNAYKEE--------LPQFIIDR 283
RVP+LD F++ A++IDP+EK GR K +LR A+ LPQ I+ R
Sbjct 408 RVPFLDKDFLEVALNIDPEEKMYINGRKEKYILRKAFDTTHDSSLQPYLPQDILWR 463
> At5g10240
Length=578
Score = 236 bits (601), Expect = 6e-62, Method: Compositional matrix adjust.
Identities = 126/285 (44%), Positives = 179/285 (62%), Gaps = 16/285 (5%)
Query 5 FPPGHSFYFKSSKASGIMERFYKPQWMQPEAPIPSAACDLSRLREELEKAVSKRMMCDVP 64
FPPGH + SSK G + R+Y P W +PS D +R EKAV KR+M DVP
Sbjct 174 FPPGHIY---SSKQGG-LRRWYNPPWFS--EVVPSTPYDPLVVRNTFEKAVIKRLMTDVP 227
Query 65 FGVLLYGGIDSAIIASIVCRLYNERFNKEQNEHFWTPKIHSFAIGLKGSRDLECSRMVAK 124
FGVLL GG+DS+++AS+ R K + W K+H+F IGLKGS DL+ R VA
Sbjct 228 FGVLLSGGLDSSLVASVALR----HLEKSEAACQWGSKLHTFCIGLKGSPDLKAGREVAD 283
Query 125 HVGCTHHEFTFEAQEAIDALSDLIYMIESYDVLTVRAALLNYFLYRLIKSIGVKMVITGE 184
++G HHE F Q+ IDA+ ++IY +E+YDV T+RA+ + + R IKS+GVKMV++GE
Sbjct 284 YLGTRHHELHFTVQDGIDAIEEVIYHVETYDVTTIRASTPMFLMSRKIKSLGVKMVLSGE 343
Query 185 GAADLFGCAGDMQEEQDPEALHKAIVKRMSDMHFTDSLRANKASMCWGVEARVPYLDVSF 244
G+ ++FG + + + H+ +++ +H D LRANK++ WGVEARVP+LD F
Sbjct 344 GSDEIFGGYLYFHKAPNKKEFHEETCRKIKALHQYDCLRANKSTSAWGVEARVPFLDKEF 403
Query 245 VDYAMSIDPKEK--RCSGGRLPKQLLRNAYKEE----LPQFIIDR 283
++ AMSIDP+ K R GR+ K +LRNA+ +E LP+ I+ R
Sbjct 404 INVAMSIDPEWKMIRPDLGRIEKWVLRNAFDDEKNPYLPKHILYR 448
> At5g65010
Length=578
Score = 234 bits (597), Expect = 1e-61, Method: Compositional matrix adjust.
Identities = 124/285 (43%), Positives = 180/285 (63%), Gaps = 16/285 (5%)
Query 5 FPPGHSFYFKSSKASGIMERFYKPQWMQPEAPIPSAACDLSRLREELEKAVSKRMMCDVP 64
FPPGH + SSK G + R+Y P W + +PS D LR EKAV KR+M DVP
Sbjct 174 FPPGHIY---SSKQGG-LRRWYNPPWYNEQ--VPSTPYDPLVLRNAFEKAVIKRLMTDVP 227
Query 65 FGVLLYGGIDSAIIASIVCRLYNERFNKEQNEHFWTPKIHSFAIGLKGSRDLECSRMVAK 124
FGVLL GG+DS+++A++ R K + W ++H+F IGL+GS DL+ R VA
Sbjct 228 FGVLLSGGLDSSLVAAVALR----HLEKSEAARQWGSQLHTFCIGLQGSPDLKAGREVAD 283
Query 125 HVGCTHHEFTFEAQEAIDALSDLIYMIESYDVLTVRAALLNYFLYRLIKSIGVKMVITGE 184
++G HHEF F Q+ IDA+ ++IY IE+YDV T+RA+ + + R IKS+GVKMV++GE
Sbjct 284 YLGTRHHEFQFTVQDGIDAIEEVIYHIETYDVTTIRASTPMFLMSRKIKSLGVKMVLSGE 343
Query 185 GAADLFGCAGDMQEEQDPEALHKAIVKRMSDMHFTDSLRANKASMCWGVEARVPYLDVSF 244
G+ ++ G + + + H+ +++ +H D LRANK++ WGVEARVP+LD F
Sbjct 344 GSDEILGGYLYFHKAPNKKEFHEETCRKIKALHQFDCLRANKSTSAWGVEARVPFLDKEF 403
Query 245 VDYAMSIDPKEK--RCSGGRLPKQLLRNAYKEE----LPQFIIDR 283
++ AMSIDP+ K + GR+ K +LRNA+ +E LP+ I+ R
Sbjct 404 LNVAMSIDPEWKLIKPDLGRIEKWVLRNAFDDEERPYLPKHILYR 448
> YGR124w
Length=572
Score = 232 bits (591), Expect = 8e-61, Method: Compositional matrix adjust.
Identities = 128/291 (43%), Positives = 179/291 (61%), Gaps = 31/291 (10%)
Query 5 FPPGHSFYFKSSKASGIMERFYKPQWMQPEAPIPSAACDLSRLREELEKAVSKRMMCDVP 64
FPPGH + ++ K + R++ P W+ E IPS D +R LEKAV KR+M +VP
Sbjct 175 FPPGHVYDSETDK----ITRYFTPDWLD-EKRIPSTPVDYHAIRHSLEKAVRKRLMAEVP 229
Query 65 FGVLLYGGIDSAIIASIVCRLYNERFNKEQNE----------------HFWTP---KIHS 105
+GVLL GG+DS++IA+I R E+ N + NE H T ++HS
Sbjct 230 YGVLLSGGLDSSLIAAIAAR-ETEKANADANEDNNVDEKQLAGIDDQGHLHTSGWSRLHS 288
Query 106 FAIGLKGSRDLECSRMVAKHVGCTHHEFTFEAQEAIDALSDLIYMIESYDVLTVRAALLN 165
FAIGL + DL+ +R VAK +G HHE TF QE +DAL D+IY +E+YDV T+RA+
Sbjct 289 FAIGLPNAPDLQAARKVAKFIGSIHHEHTFTLQEGLDALDDVIYHLETYDVTTIRASTPM 348
Query 166 YFLYRLIKSIGVKMVITGEGAADLFGCAGDMQEEQDPEA--LHKAIVKRMSDMHFTDSLR 223
+ L R IK+ GVKMV++GEG+ ++FG G + Q P A H V+R+ ++H D LR
Sbjct 349 FLLSRKIKAQGVKMVLSGEGSDEIFG--GYLYFAQAPSAAEFHTESVQRVKNLHLADCLR 406
Query 224 ANKASMCWGVEARVPYLDVSFVDYAMSIDPKEK--RCSGGRLPKQLLRNAY 272
ANK++M WG+EARVP+LD F+ M+IDP EK + GR+ K +LR A+
Sbjct 407 ANKSTMAWGLEARVPFLDKDFLQLCMNIDPNEKMIKPKEGRIEKYILRKAF 457
> YPR145w
Length=572
Score = 231 bits (588), Expect = 2e-60, Method: Compositional matrix adjust.
Identities = 127/295 (43%), Positives = 177/295 (60%), Gaps = 32/295 (10%)
Query 2 VREFPPGHSFYFKSSKASGIMERFYKPQWMQPEAPIPSAACDLSRLREELEKAVSKRMMC 61
+ FPPGH + K+ K + R++ P W+ E IPS D +R LEKAV KR+M
Sbjct 172 ITAFPPGHVYDSKTDK----ITRYFTPDWLD-EKRIPSTPIDYMAIRHSLEKAVRKRLMA 226
Query 62 DVPFGVLLYGGIDSAIIASIVCRLYNERFNKEQNEHF--------------------WTP 101
+VP+GVLL GG+DS++IASI R + N + + WT
Sbjct 227 EVPYGVLLSGGLDSSLIASIAARETAKATNDVEPSTYDSKARHLAGIDDDGKLHTAGWT- 285
Query 102 KIHSFAIGLKGSRDLECSRMVAKHVGCTHHEFTFEAQEAIDALSDLIYMIESYDVLTVRA 161
+HSFAIGL + DL+ +R VAK +G HHE TF QE +DAL D+IY +E+YDV T+RA
Sbjct 286 SLHSFAIGLPNAPDLQAARKVAKFIGSIHHEHTFTLQEGLDALDDVIYHLETYDVTTIRA 345
Query 162 ALLNYFLYRLIKSIGVKMVITGEGAADLFGCAGDMQEEQDPEA--LHKAIVKRMSDMHFT 219
+ + L R IK+ GVKMV++GEG+ ++FG G + Q P A H V+R+ ++H
Sbjct 346 STPMFLLSRKIKAQGVKMVLSGEGSDEIFG--GYLYFAQAPSAAEFHTESVQRVKNLHLA 403
Query 220 DSLRANKASMCWGVEARVPYLDVSFVDYAMSIDPKEK--RCSGGRLPKQLLRNAY 272
D LRANK++M WG+EARVP+LD F+ M+IDP EK + GR+ K +LR A+
Sbjct 404 DCLRANKSTMAWGLEARVPFLDREFLQLCMNIDPNEKMIKPKEGRIEKYILRKAF 458
> At3g47340
Length=584
Score = 230 bits (586), Expect = 3e-60, Method: Compositional matrix adjust.
Identities = 122/286 (42%), Positives = 182/286 (63%), Gaps = 18/286 (6%)
Query 5 FPPGHSFYFKSSKASGIMERFYKPQWMQPEAPIPSAACDLSRLREELEKAVSKRMMCDVP 64
FPPGH F SSK G +++Y P W +PS + +R E AV KR+M DVP
Sbjct 174 FPPGH---FYSSKLGGF-KQWYNPPWFNES--VPSTPYEPLAIRRAFENAVIKRLMTDVP 227
Query 65 FGVLLYGGIDSAIIASIVCR-LYNERFNKEQNEHFWTPKIHSFAIGLKGSRDLECSRMVA 123
FGVLL GG+DS+++ASI R L + K+ W P++HSF +GL+GS DL+ + VA
Sbjct 228 FGVLLSGGLDSSLVASITARHLAGTKAAKQ-----WGPQLHSFCVGLEGSPDLKAGKEVA 282
Query 124 KHVGCTHHEFTFEAQEAIDALSDLIYMIESYDVLTVRAALLNYFLYRLIKSIGVKMVITG 183
+++G HHEF F Q+ IDA+ D+IY +E+YDV T+RA+ + + R IKS+GVKMV++G
Sbjct 283 EYLGTVHHEFHFSVQDGIDAIEDVIYHVETYDVTTIRASTPMFLMSRKIKSLGVKMVLSG 342
Query 184 EGAADLFGCAGDMQEEQDPEALHKAIVKRMSDMHFTDSLRANKASMCWGVEARVPYLDVS 243
EGA ++FG + + + H+ +++ +H D LRANK++ +G+EARVP+LD
Sbjct 343 EGADEIFGGYLYFHKAPNKKEFHQETCRKIKALHKYDCLRANKSTSAFGLEARVPFLDKD 402
Query 244 FVDYAMSIDPKEK--RCSGGRLPKQLLRNAYKEE----LPQFIIDR 283
F++ AMS+DP+ K + GR+ K +LR A+ +E LP+ I+ R
Sbjct 403 FINTAMSLDPESKMIKPEEGRIEKWVLRRAFDDEERPYLPKHILYR 448
> CE04760
Length=567
Score = 182 bits (462), Expect = 8e-46, Method: Compositional matrix adjust.
Identities = 106/289 (36%), Positives = 161/289 (55%), Gaps = 22/289 (7%)
Query 1 RVREFPPGHSFYFKSSKASGIMERFYKPQWMQPEAPIPSAACDLSRLREELEKAVSKRMM 60
++ FPPGH + K+ R++ P+W I DL L++ + +V KR+M
Sbjct 175 KIESFPPGHYYTPKTG-----FVRYFNPEWFDFRKAI--HPLDLKLLQKTMIASVHKRLM 227
Query 61 CDVPFGVLLYGGIDSAIIASIVCRLYNERFNKEQNEHFWTPKIHSFAIGL-KGSRDLECS 119
D P GVLL GG+DS++++SI R R +HSF+IG+ S D+ +
Sbjct 228 SDAPIGVLLSGGLDSSLVSSIASREMKRR----------GMAVHSFSIGVDHNSPDVVAA 277
Query 120 RMVAKHVGCTHHEFTFEAQEAIDALSDLIYMIESYDVLTVRAALLNYFLYRLIKSIGVKM 179
R VAK +G THHEF F +E I L LI+ +ESYDV ++RA+ YFL I+ +G+K+
Sbjct 278 RKVAKFIGTTHHEFYFSIEEGIKNLRKLIWHLESYDVTSIRASTPMYFLSEEIRKLGIKV 337
Query 180 VITGEGAADLFGCAGDMQEEQDPEALHKAIVKRMSDMHFTDSLRANKASMCWGVEARVPY 239
V++GEGA ++FG E K + R+ ++ +D LRA+K+SM VE RVP+
Sbjct 338 VLSGEGADEIFGGYLYFHNAPSDEDFQKETIDRVLHLYTSDCLRADKSSMAHSVEVRVPF 397
Query 240 LDVSFVDYAMSIDPKEKRCS----GGRLPKQLLRNAYKEELPQFIIDRI 284
LD +FV+ A+S+DP KR G K +LR+A+ + ++ D I
Sbjct 398 LDKAFVEAAVSLDPAFKRPQKLEDGRNCEKFVLRSAFNTDQYPYLPDEI 446
> CE09642
Length=551
Score = 149 bits (377), Expect = 5e-36, Method: Compositional matrix adjust.
Identities = 95/293 (32%), Positives = 156/293 (53%), Gaps = 33/293 (11%)
Query 1 RVREFPPGHSFYFKSSKASGIMERFYKPQWMQPEA--PIPSAA-------CDLSRLREEL 51
RV FPPG + F + +QP+ +P+ A C + +R+ L
Sbjct 167 RVEYFPPG---------CCATISLFAPTRPVQPQQYYSVPTIADRFLSIDCTKTLVRDVL 217
Query 52 EKAVSKRMMCDVPFGVLLYGGIDSAIIASIVCRLYNERFNKEQNEHFWTPKIHSFAIGLK 111
K+V KR+M + FG +L GG+DS++IASI R F K +F++G +
Sbjct 218 VKSVEKRLMGNRNFGFMLSGGLDSSLIASIATR-------------FLKQKPIAFSVGFE 264
Query 112 GSRDLECSRMVAKHVGCTHHEFTFEAQEAIDALSDLIYMIESYDVLTVRAALLNYFLYRL 171
S DLE ++ VA ++ H Q+ ID + ++++ +E++D L +R + +Y L +
Sbjct 265 DSPDLENAKKVADYLKIPHEVLVITPQQCIDIIPEVVFALETFDPLIIRCGIAHYLLCQH 324
Query 172 I-KSIGVKMVITGEGAADLFGCAGDMQEEQDPEALHKAIVKRMSDMHFTDSLRANKASMC 230
I KS VK++++GEGA +LFG MQ + LHK I++RM +H D LR ++++ C
Sbjct 325 ISKSSDVKVLLSGEGADELFGSYAYMQRAPNALHLHKEILRRMHHLHQYDVLRCDRSTSC 384
Query 231 WGVEARVPYLDVSFVDYAMSIDPKEKRCSGGRLPKQLLRNAYKEELPQFIIDR 283
G+E RVP+LD F+D + P K +L K +LR+A++ LP ++ R
Sbjct 385 HGLEIRVPFLDKRFIDLVSRLPPSYKLMP-MKLEKHVLRSAFEGWLPDEVLWR 436
> Hs19718772
Length=561
Score = 126 bits (317), Expect = 5e-29, Method: Compositional matrix adjust.
Identities = 77/242 (31%), Positives = 136/242 (56%), Gaps = 12/242 (4%)
Query 45 SRLREELEKAVSKRMMCDVPFGVLLYGGIDSAIIASIVCRLYNERFNKEQNEHFWTPKIH 104
+ LR AV KR+M D G LL GG+DS+++A+ + + + + Q ++ +
Sbjct 233 NNLRILFNNAVKKRLMTDRRIGCLLSGGLDSSLVAATLLK----QLKEAQVQY----PLQ 284
Query 105 SFAIGLKGSRDLECSRMVAKHVGCTHHEFTFEAQEAIDALSDLIYMIESYDVLTVRAALL 164
+FAIG++ S DL +R VA H+G H+E F ++E I AL ++I+ +E+YD+ TVRA++
Sbjct 285 TFAIGMEDSPDLLAARKVADHIGSEHYEVLFNSEEGIQALDEVIFSLETYDITTVRASVG 344
Query 165 NYFLYRLI-KSIGVKMVITGEGAADLFGCAGDMQEEQDPEALHKAIVKRMSDMHFTDSLR 223
Y + + I K+ ++ +GEG+ +L + PE + + + +++ D LR
Sbjct 345 MYLISKYIRKNTDSVVIFSGEGSDELTQGYIYFHKAPSPEKAEEESERLLRELYLFDVLR 404
Query 224 ANKASMCWGVEARVPYLDVSFVDYAMSIDPKEKRCSGGRLPKQLLRNAYKEE--LPQFII 281
A++ + G+E RVP+LD F Y +S+ P E R + K LLR +++ +P+ I+
Sbjct 405 ADRTTAAHGLELRVPFLDHRFSSYYLSL-PPEMRIPKNGIEKHLLRETFEDSNLIPKEIL 463
Query 282 DR 283
R
Sbjct 464 WR 465
> Hs20539333
Length=430
Score = 82.8 bits (203), Expect = 8e-16, Method: Compositional matrix adjust.
Identities = 58/233 (24%), Positives = 115/233 (49%), Gaps = 37/233 (15%)
Query 45 SRLREELEKAVSKRMMCD-VPFGVLLYGGIDSAIIASIVCRLYNERFNKEQNEHFWTPKI 103
++LR A+ +R M D +PF G+ ++A+ + + + + Q ++ +
Sbjct 155 NKLRILFHDAIKRRFMIDRLPF----IRGLGLQLVAATLLK----QLKEAQVQY----PL 202
Query 104 HSFAIGLKGSRDLECSRMVAKHVGCTHHEFTFEAQEAIDALSDLIYMIESYDVLTVRAAL 163
+F IG++ S DL +R VA H+G HHE F ++E AL ++I+ +E+Y++ T A++
Sbjct 203 QTFVIGMEDSPDLLAARKVANHIGSEHHEVLFNSEEGFQALDEVIFSLETYNITTADASV 262
Query 164 LNYFLYRLI-KSIGVKMVITGEGAADLFGC----------AGDMQEEQDPEALHKAIVKR 212
Y + + I K+ ++ +GEG+ +L A + +++ E L
Sbjct 263 SMYLISKYIRKNTDSVVIFSGEGSDELIQAKLMVLLFVKWAPSEKAKEESERL------- 315
Query 213 MSDMHFTDSLRANKASMCWGVEARVPYLDVSFVDYAMSIDPKEKRCSGGRLPK 265
+ +++ D L A++ + +E RVP+LD F Y + + + R+PK
Sbjct 316 LRELYLFDVLHADQTTAAHSLELRVPFLDHQFSSYYLYL------STEMRIPK 362
> YML096w
Length=525
Score = 32.3 bits (72), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 49/228 (21%), Positives = 96/228 (42%), Gaps = 40/228 (17%)
Query 47 LREELEK---AVSKRMMCDVPFGVLLYGGIDSAIIASIVCRLYNERFNK-------EQNE 96
LR+ ++K ++ R + + P VL GGID ++I +++C + E K N
Sbjct 234 LRDSVKKRVESIHPRHIENSPIAVLFSGGIDCSVIVALICEVLQENDYKCGKPVIELLNV 293
Query 97 HFWTPKIHSFAIGLKGSRDLECSRMVAKHVGCTHHEFTFEAQEAIDALSD---------- 146
F P+ F + D + S AK + + + E +D D
Sbjct 294 SFENPRTGLFP---SDTPDRKLSINSAKTLQNLYPNVDIKLVE-VDVPYDEYLKWKPFVI 349
Query 147 -LIYMIESYDVLTVRAALLNYFLYR---LIKSIGVK---------MVITGEGAADLFGCA 193
L+Y ++ L++ A +F R + S+ + ++ +G GA +L+G
Sbjct 350 NLMYPKQTEMDLSIAIAF--FFASRGRGFLTSLNGERTPYQRHGIVLFSGLGADELYGGY 407
Query 194 GDMQEEQDPEALHKAIVKRMSDMHFTDSLRANKASMCWGVEARVPYLD 241
+ P L + + +++++++ + R +K GVE R P+LD
Sbjct 408 HKFA-NKPPHELVEELTRQINNIYDRNLNRDDKVIAHNGVEVRYPFLD 454
> 7292212
Length=2086
Score = 32.3 bits (72), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 32/152 (21%), Positives = 57/152 (37%), Gaps = 18/152 (11%)
Query 26 YKPQWMQPE--APIPSAACDLSRLREELEKAVSKRMMCDVPFGVLLYGGIDSAIIASIVC 83
YK W++ +PI LSR + + K + +P +++ + ++V
Sbjct 588 YKKLWLRGRRLSPITQPTFFLSRWFIKASDLIEKSVFVRIPLAIMVLFLLYVMSSETVVS 647
Query 84 RLYNERFNKEQNEHFWTPKIHSFAIGLKGSRDLECSRMVAKHVGCTH--HEFTFEAQEAI 141
F+ P I +GL ++ HV H + F FE +
Sbjct 648 PFRTSEFHNLMLVIIGIPLIIKLGVGLA---------ILGSHVITVHAYYGFAFERSQTT 698
Query 142 D-----ALSDLIYMIESYDVLTVRAALLNYFL 168
+ +L+ YM+ + V+ VR LNY L
Sbjct 699 NIGVTSSLAHSWYMVAFFIVVVVREGYLNYIL 730
> 7303694
Length=949
Score = 30.8 bits (68), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 28/51 (54%), Gaps = 0/51 (0%)
Query 195 DMQEEQDPEALHKAIVKRMSDMHFTDSLRANKASMCWGVEARVPYLDVSFV 245
D+ E P+A+H+++ ++ SL+ N+ ++ V RVPY DV V
Sbjct 763 DLSLEVTPKAVHESLKQQNYTKALVMSLKLNEPNLIALVLERVPYKDVELV 813
> 7295991
Length=785
Score = 30.4 bits (67), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 25/49 (51%), Gaps = 2/49 (4%)
Query 133 FTFEAQEAIDALSDLIYMIESYDVLTVRAALLNYFLYRLIKSIGVKMVI 181
T E+Q+ ID L YM S+DV TV +NYF L + ++ I
Sbjct 655 LTGESQDGIDILQS--YMDTSFDVQTVALVAINYFRQELFEDKRIQYWI 701
> 7289856
Length=564
Score = 30.0 bits (66), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 27/106 (25%), Positives = 47/106 (44%), Gaps = 16/106 (15%)
Query 178 KMVITGEGAADLFG-----------CAGDMQEEQDPEALHKAIVKRMSDMHFTDSLRANK 226
++ + G GA +LFG C G+ E Q A+ + + + + R ++
Sbjct 422 RVALIGSGADELFGGYTRYRNSYSRCLGNDLERQ--LAVQNELERDWQRISARNLARDDR 479
Query 227 ASMCWGVEARVPYLDVSFVDYAMSIDPKEKRCSG---GRLPKQLLR 269
G AR P+++ +FV + S++ +K C G G K LLR
Sbjct 480 IIADTGKTARSPFIEENFVKFIRSLEVYQKCCFGFPEGVGDKLLLR 525
> 7293513
Length=218
Score = 30.0 bits (66), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 14/48 (29%), Positives = 27/48 (56%), Gaps = 0/48 (0%)
Query 106 FAIGLKGSRDLECSRMVAKHVGCTHHEFTFEAQEAIDALSDLIYMIES 153
++IG G D++C+ + +V CT T + + AI++ S+ + ES
Sbjct 45 YSIGTVGYTDVDCNFIDFTYVCCTSDSLTHKVKRAINSFSEKLRSNES 92
Lambda K H
0.323 0.137 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6410666326
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40