bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1048_orf1
Length=178
Score E
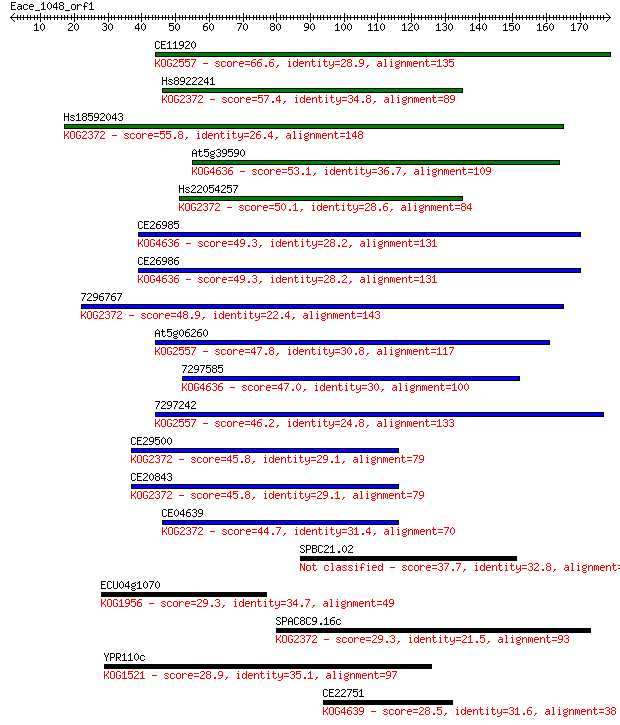
Sequences producing significant alignments: (Bits) Value
CE11920 66.6 3e-11
Hs8922241 57.4 1e-08
Hs18592043 55.8 5e-08
At5g39590 53.1 3e-07
Hs22054257 50.1 2e-06
CE26985 49.3 4e-06
CE26986 49.3 5e-06
7296767 48.9 6e-06
At5g06260 47.8 1e-05
7297585 47.0 2e-05
7297242 46.2 4e-05
CE29500 45.8 4e-05
CE20843 45.8 4e-05
CE04639 44.7 1e-04
SPBC21.02 37.7 0.012
ECU04g1070 29.3 4.0
SPAC8C9.16c 29.3 4.0
YPR110c 28.9 6.3
CE22751 28.5 7.0
> CE11920
Length=399
Score = 66.6 bits (161), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 39/135 (28%), Positives = 64/135 (47%), Gaps = 2/135 (1%)
Query 44 FPFPPKAPWTCLYASWKHGTSFSRLCANCFFYSAPMVAVVKTEEGQVLGAIISCELKEGG 103
PF + WT LY++ KHG SFS+L C P + V+++ +G+ G S G
Sbjct 192 LPFDRRKNWTLLYSNMKHGQSFSQL-VKCINGEGPCMIVIRSMKGRRFGFFASQGFL-AG 249
Query 104 HNFFGDANTCLFSLEPQLNILRTSGLGRIFIYLHTSNKFHPIGMGFGGQVGAFRFWIGDE 163
+ G A LF L P++ +G ++YL+ + HP G+G GG + +I +E
Sbjct 250 PQYRGTAECFLFQLAPKIATFDATGRTENYVYLNYQQQQHPNGLGIGGTESVWPLFIHEE 309
Query 164 MKDCYITKSDCTYSP 178
K+ ++ P
Sbjct 310 FGGGTCQKNSSSFEP 324
> Hs8922241
Length=269
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 31/92 (33%), Positives = 46/92 (50%), Gaps = 4/92 (4%)
Query 46 FPPKA---PWTCLYASWKHGTSFSRLCANCFFYSAPMVAVVKTEEGQVLGAIISCELKEG 102
PP+ PWT +Y + KHGTS L P++ V+K +GQV GA+ S LK
Sbjct 122 LPPRTIGYPWTLVYGTGKHGTSLKTLYRTMTGLDTPVLMVIKDSDGQVFGALASEPLK-V 180
Query 103 GHNFFGDANTCLFSLEPQLNILRTSGLGRIFI 134
F+G T +F+ P+ + + +G FI
Sbjct 181 SDGFYGTGETFVFTFCPEFEVFKWTGDDMFFI 212
> Hs18592043
Length=197
Score = 55.8 bits (133), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 39/151 (25%), Positives = 67/151 (44%), Gaps = 15/151 (9%)
Query 17 HGQGPLDARSKCFTDEHAFVLRMASTLFPFPPKA---PWTCLYASWKHGTSFSRLCANCF 73
HG+ D R + + +L F FPP+ PW+ ++ + + G S L
Sbjct 26 HGRLSRDRRMRGLRWRYTRLLS-----FHFPPRVTGHPWSLVFCTSRDGFSLQSLYRRME 80
Query 74 FYSAPMVAVVKTEEGQVLGAIISCELKEGGHNFFGDANTCLFSLEPQLNILRTSGLGRIF 133
S P++ V++ ++GQ+ GA S ++ F+G T LFS PQL + + +G F
Sbjct 81 GCSGPVLLVLRDQDGQIFGAFSSSAIRL-SKGFYGTGETFLFSFSPQLKVFKWTGSNSFF 139
Query 134 IYLHTSNKFHPIGMGFGGQVGAFRFWIGDEM 164
+ + MG G G F W+ ++
Sbjct 140 V----KGDLDSLMMGSGS--GRFGLWLDGDL 164
> At5g39590
Length=542
Score = 53.1 bits (126), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 40/130 (30%), Positives = 65/130 (50%), Gaps = 26/130 (20%)
Query 55 LYASWKHGTSFSRLCANCFFYSAPMVAVV-----------KTEEGQVLGAIISCELKEGG 103
LY S+ HG +RL +N Y AP++ ++ +E V+GAI L++G
Sbjct 320 LYRSYYHGKGMNRLWSNVEGYHAPILVIISASCKVEHEATSSERKWVIGAI----LQQGF 375
Query 104 HN---FFGDANTCLFSLEPQLNILRTSGLGRIFIY--LHTSNKFH-----PIGMGFGGQV 153
N F+G + LFS+ P + +SG + F Y LH + + P+G+GFGG +
Sbjct 376 ENRDAFYGSSGN-LFSISPVFHAFSSSGKEKNFAYSHLHPAGGVYDAHPKPVGIGFGGTL 434
Query 154 GAFRFWIGDE 163
G R +I ++
Sbjct 435 GNERIFIDED 444
> Hs22054257
Length=942
Score = 50.1 bits (118), Expect = 2e-06, Method: Composition-based stats.
Identities = 24/84 (28%), Positives = 40/84 (47%), Gaps = 1/84 (1%)
Query 51 PWTCLYASWKHGTSFSRLCANCFFYSAPMVAVVKTEEGQVLGAIISCELKEGGHNFFGDA 110
PW Y++ +HGTS L +P++ V+K + Q+ GA + K H ++G
Sbjct 803 PWRLAYSTLEHGTSLKTLYRKSASLDSPVLLVIKDMDNQIFGAYATHPFKFSDH-YYGTG 861
Query 111 NTCLFSLEPQLNILRTSGLGRIFI 134
T L++ P + + SG FI
Sbjct 862 ETFLYTFSPHFKVFKWSGENSYFI 885
> CE26985
Length=415
Score = 49.3 bits (116), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 37/136 (27%), Positives = 61/136 (44%), Gaps = 12/136 (8%)
Query 39 MASTLFPFPP-----KAPWTCLYASWKHGTSFSRLCANCFFYSAPMVAVVKTEEGQVLGA 93
+ + FP P ++ WT LY S +HG S +R F Y P V + + ++G+V+
Sbjct 204 LPAVYFPAKPTETSGESHWTPLYTSLQHGISTNRFETLVFDYRGPTVTIFRMKDGRVVVI 263
Query 94 IISCELKEGGHNFFGDANTCLFSLEPQLNILRTSGLGRIFIYLHTSNKFHPIGMGFGGQV 153
E + G N FG T F + P NI R G I+ L + + G+ F ++
Sbjct 264 AADQEWRHSG-NRFGGTFTSFFEIVP--NIRRIDGANSIYCNLKLRSSAY--GLSFKNEL 318
Query 154 GAFRFWIGDEMKDCYI 169
+ + DE+ D +
Sbjct 319 KIEKDF--DEILDIEV 332
> CE26986
Length=411
Score = 49.3 bits (116), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 37/136 (27%), Positives = 61/136 (44%), Gaps = 12/136 (8%)
Query 39 MASTLFPFPP-----KAPWTCLYASWKHGTSFSRLCANCFFYSAPMVAVVKTEEGQVLGA 93
+ + FP P ++ WT LY S +HG S +R F Y P V + + ++G+V+
Sbjct 200 LPAVYFPAKPTETSGESHWTPLYTSLQHGISTNRFETLVFDYRGPTVTIFRMKDGRVVVI 259
Query 94 IISCELKEGGHNFFGDANTCLFSLEPQLNILRTSGLGRIFIYLHTSNKFHPIGMGFGGQV 153
E + G N FG T F + P NI R G I+ L + + G+ F ++
Sbjct 260 AADQEWRHSG-NRFGGTFTSFFEIVP--NIRRIDGANSIYCNLKLRSSAY--GLSFKNEL 314
Query 154 GAFRFWIGDEMKDCYI 169
+ + DE+ D +
Sbjct 315 KIEKDF--DEILDIEV 328
> 7296767
Length=1325
Score = 48.9 bits (115), Expect = 6e-06, Method: Composition-based stats.
Identities = 32/143 (22%), Positives = 62/143 (43%), Gaps = 9/143 (6%)
Query 22 LDARSKCFTDEHAFVLRMASTLFPFPPKAPWTCLYASWKHGTSFSRLCANCFFYSAPMVA 81
L +++ T+EH ++ S L W+ ++++ +HG + + L +P++
Sbjct 1159 LIGKTEILTEEHR--EKLCSHLPARAEGYSWSLIFSTSQHGFALNSLYRKMARLESPVLI 1216
Query 82 VVKTEEGQVLGAIISCELKEGGHNFFGDANTCLFSLEPQLNILRTSGLGRIFIYLHTSNK 141
V++ E V GA+ SC L H F+G + L+ P + +G FI K
Sbjct 1217 VIEDTEHNVFGALTSCSLHVSDH-FYGTGESLLYKFNPSFKVFHWTGENMYFI------K 1269
Query 142 FHPIGMGFGGQVGAFRFWIGDEM 164
+ + G G F W+ ++
Sbjct 1270 GNMESLSIGAGDGRFGLWLDGDL 1292
> At5g06260
Length=424
Score = 47.8 bits (112), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 36/120 (30%), Positives = 55/120 (45%), Gaps = 4/120 (3%)
Query 44 FPFPPKAPWTCLYASWKHGTSFSRLCANCFFYS-APMVAVVKTEEGQVLGAIISCELKEG 102
P W LY S HG SF+ + + V ++K EG V G S + E
Sbjct 230 LPHHELVEWKLLYHSSVHGQSFNTFLGHTSNTGMSASVLIIKDTEGYVYGGYAS-QPWER 288
Query 103 GHNFFGDANTCLFSLEPQLNILRTSGLGRIFIYLHT--SNKFHPIGMGFGGQVGAFRFWI 160
+F+GD + LF L P+ I R +G + T +++ P G+GFGG++ F +I
Sbjct 289 YSDFYGDMKSFLFQLNPKAAIYRPTGANTNIQWCATNFTSENIPNGIGFGGKINHFGLFI 348
> 7297585
Length=526
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 30/100 (30%), Positives = 46/100 (46%), Gaps = 4/100 (4%)
Query 52 WTCLYASWKHGTSFSRLCANCFFYSAPMVAVVKTEEGQVLGAIISCELKEGGHNFFGDAN 111
WT LY S +HG +R + Y P + ++ T++ Q E KE H F G
Sbjct 346 WTLLYNSNEHGVGANRFLHHVLGYRGPTLVLLHTKDEQTYCVASPSEWKE-THLFVGGEG 404
Query 112 TCLFSLEPQLNILRTSGLGRIFIYLHTSNKFHPIGMGFGG 151
+C+ L P+ IL +YL+TS + +P G+ G
Sbjct 405 SCVIQLLPKFVILEKKP---NILYLNTSIRGYPKGLRAGA 441
> 7297242
Length=448
Score = 46.2 bits (108), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 33/133 (24%), Positives = 56/133 (42%), Gaps = 2/133 (1%)
Query 44 FPFPPKAPWTCLYASWKHGTSFSRLCANCFFYSAPMVAVVKTEEGQVLGAIISCELKEGG 103
P + W L++S +G SFS + P + ++ E+ + G S E
Sbjct 247 LPREHRHKWRFLFSSKINGESFSTMLGKVLD-KGPTLFFIEDEDQYIFGGYAS-ETWSVK 304
Query 104 HNFFGDANTCLFSLEPQLNILRTSGLGRIFIYLHTSNKFHPIGMGFGGQVGAFRFWIGDE 163
F GD ++ L++L P + + + YL+ + + P G+G GGQ + WI
Sbjct 305 PQFGGDDSSLLYTLSPAMRCFSATTYNNHYQYLNLNQQTMPNGLGMGGQFDFWGLWIDCS 364
Query 164 MKDCYITKSDCTY 176
D +S TY
Sbjct 365 FGDGQSVESCTTY 377
> CE29500
Length=817
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 23/85 (27%), Positives = 43/85 (50%), Gaps = 7/85 (8%)
Query 37 LRMASTLFPFPPKA---PWTCLYASWKHGTSFSRLCANCFFYS---APMVAVVKTEEGQV 90
L + + PP+A PW +Y S KHG S + + + +P++ +++ + V
Sbjct 649 LMIRQVMDILPPRAEGYPWVNIYNSEKHGFSLATMYRKMAEFDEDLSPVLLIIRDTKEHV 708
Query 91 LGAIISCELKEGGHNFFGDANTCLF 115
GA++S ++ H FFG ++CL
Sbjct 709 FGAVVSSAIRPNDH-FFGTGDSCLL 732
> CE20843
Length=838
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 23/85 (27%), Positives = 43/85 (50%), Gaps = 7/85 (8%)
Query 37 LRMASTLFPFPPKA---PWTCLYASWKHGTSFSRLCANCFFYS---APMVAVVKTEEGQV 90
L + + PP+A PW +Y S KHG S + + + +P++ +++ + V
Sbjct 670 LMIRQVMDILPPRAEGYPWVNIYNSEKHGFSLATMYRKMAEFDEDLSPVLLIIRDTKEHV 729
Query 91 LGAIISCELKEGGHNFFGDANTCLF 115
GA++S ++ H FFG ++CL
Sbjct 730 FGAVVSSAIRPNDH-FFGTGDSCLL 753
> CE04639
Length=536
Score = 44.7 bits (104), Expect = 1e-04, Method: Composition-based stats.
Identities = 22/76 (28%), Positives = 40/76 (52%), Gaps = 7/76 (9%)
Query 46 FPPKA---PWTCLYASWKHGTSFSRLCANCFFYS---APMVAVVKTEEGQVLGAIISCEL 99
PP+A PW +Y S KHG S + + + +P++ +++ + V GA++S +
Sbjct 377 LPPRAEGYPWVNIYNSEKHGFSLATMYRKMAEFDEDLSPVLLIIRDTKEHVFGAVVSSAI 436
Query 100 KEGGHNFFGDANTCLF 115
+ H FFG ++CL
Sbjct 437 RPNDH-FFGTGDSCLL 451
> SPBC21.02
Length=511
Score = 37.7 bits (86), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 21/64 (32%), Positives = 32/64 (50%), Gaps = 5/64 (7%)
Query 87 EGQVLGAIISCELKEGGHNFFGDANTCLFSLEPQLNILRTSGLGRIFIYLHTSNKFHPIG 146
E +LGA IS ++ FFGD +T LF L+P + S L + + + +G
Sbjct 377 ENILLGAYISTRWRQSHMGFFGDHSTLLFQLQPIHQVYYASNLDKNYCMFDKN-----VG 431
Query 147 MGFG 150
+GFG
Sbjct 432 LGFG 435
> ECU04g1070
Length=621
Score = 29.3 bits (64), Expect = 4.0, Method: Composition-based stats.
Identities = 17/59 (28%), Positives = 22/59 (37%), Gaps = 10/59 (16%)
Query 28 CFTDEHAFVLRMASTLFPFPPKAPWTCLYA----------SWKHGTSFSRLCANCFFYS 76
C FV+ A F P+ WT + +W+ G F R C CFF S
Sbjct 99 CQVPTLGFVVERARERESFVPEIFWTLKFKAMHKGFKDEFTWRRGPVFDRNCVVCFFNS 157
> SPAC8C9.16c
Length=188
Score = 29.3 bits (64), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 20/93 (21%), Positives = 37/93 (39%), Gaps = 5/93 (5%)
Query 80 VAVVKTEEGQVLGAIISCELKEGGHNFFGDANTCLFSLEPQLNILRTSGLGRIFIYLHTS 139
+ V+ +G V G I L H +FG T L+ P + +G + +
Sbjct 72 ILAVRDTDGDVFGVFIPDYLIPAPH-YFGSEETFLWKYFPPKKYVHYPFVGNSNFVAYCT 130
Query 140 NKFHPIGMGFGGQVGAFRFWIGDEMKDCYITKS 172
F + FGG G + W+ ++ Y +++
Sbjct 131 KSF----LAFGGGNGRYSLWLDGSLEYAYSSRT 159
> YPR110c
Length=335
Score = 28.9 bits (63), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 34/116 (29%), Positives = 47/116 (40%), Gaps = 23/116 (19%)
Query 29 FTDEHAFVLRMAS--TLFPFPPKAPWTC--------LYA-------SWKHGTSFSRLCAN 71
FTDE+ VL + T P PK +YA + T+F A+
Sbjct 126 FTDENTIVLSLNVKCTRNPDAPKGSTDPKELYNNAHVYARDLKFEPQGRQSTTF----AD 181
Query 72 CFFYSA-PMVAVVKTEEGQVLGAIISCELKEGG-HNFFGDANTCLFSLEPQLNILR 125
C A P + + K GQ + C L GG H F +T + L PQ+NIL+
Sbjct 182 CPVVPADPDILLAKLRPGQEISLKAHCILGIGGDHAKFSPVSTASYRLLPQINILQ 237
> CE22751
Length=158
Score = 28.5 bits (62), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 12/38 (31%), Positives = 22/38 (57%), Gaps = 0/38 (0%)
Query 94 IISCELKEGGHNFFGDANTCLFSLEPQLNILRTSGLGR 131
+I + EGG+ +FG C+ ++ Q +L+T +GR
Sbjct 61 VIVLRVAEGGNKYFGAILPCVHKIDKQPMVLQTIFIGR 98
Lambda K H
0.324 0.139 0.461
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2779358582
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40