bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1065_orf1
Length=253
Score E
Sequences producing significant alignments: (Bits) Value
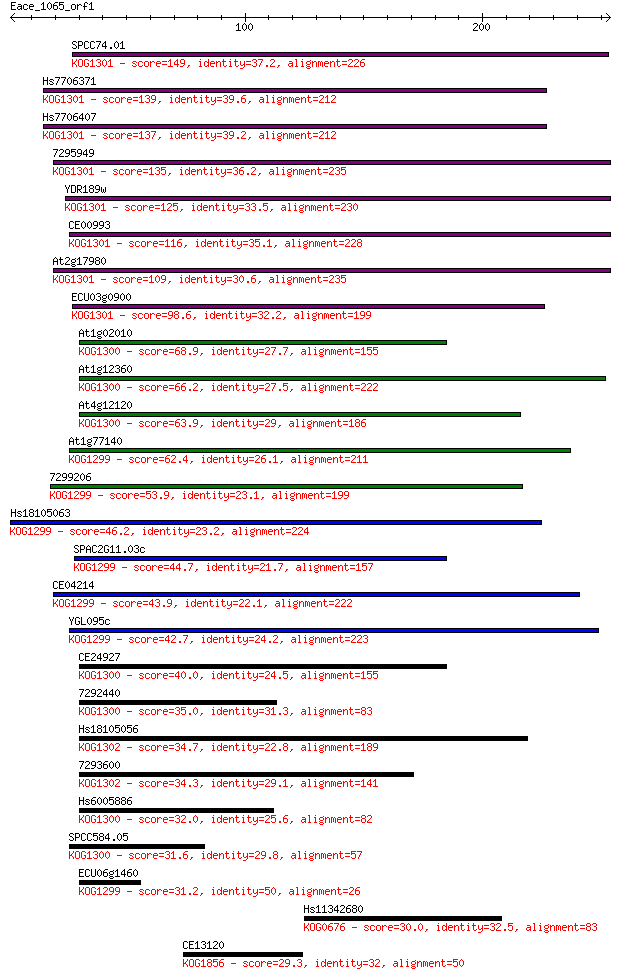
SPCC74.01 149 6e-36
Hs7706371 139 6e-33
Hs7706407 137 2e-32
7295949 135 1e-31
YDR189w 125 9e-29
CE00993 116 4e-26
At2g17980 109 5e-24
ECU03g0900 98.6 1e-20
At1g02010 68.9 1e-11
At1g12360 66.2 6e-11
At4g12120 63.9 3e-10
At1g77140 62.4 1e-09
7299206 53.9 3e-07
Hs18105063 46.2 7e-05
SPAC2G11.03c 44.7 2e-04
CE04214 43.9 3e-04
YGL095c 42.7 8e-04
CE24927 40.0 0.004
7292440 35.0 0.15
Hs18105056 34.7 0.17
7293600 34.3 0.26
Hs6005886 32.0 1.4
SPCC584.05 31.6 1.5
ECU06g1460 31.2 1.9
Hs11342680 30.0 5.1
CE13120 29.3 8.3
> SPCC74.01
Length=639
Score = 149 bits (376), Expect = 6e-36, Method: Compositional matrix adjust.
Identities = 84/235 (35%), Positives = 129/235 (54%), Gaps = 20/235 (8%)
Query 27 KPLLILIDRDFDLLTMLQHTWLYGALIHDLLQLRLNRVTVVTPGEGGKPSTTKTFDLEKR 86
+P+LIL+DR DL+ M+ H+W Y ALIHD L ++LNR+TV + +G T + +DL+
Sbjct 247 RPILILLDRTVDLIPMINHSWTYQALIHDTLNMQLNRITVESVDDGKM--TKRFYDLDGN 304
Query 87 DVFWREHAFSPFPVAAAAVSQMLTEYNAELAKVGQRPP---------DAFAQPEGDLSAD 137
D FW +A PFP A + + LT Y + +++ ++ DAFA S
Sbjct 305 DFFWESNASKPFPKVAENIDEELTRYKNDASEITRKSGVSSLEEVNVDAFAD-----STY 359
Query 138 ILKAVDVLPDLADKKRSLDAHTTIATALVSAIKDRGLDRYFEIEQSLPQDRETVVTSAVQ 197
+ AV +LP+L +K+ LD H IATAL+ AI++R LD +F++E ++ + SA+
Sbjct 360 LKSAVSLLPELTARKQILDMHMNIATALLKAIQERHLDDFFQLEDNIT----GLNRSAIL 415
Query 198 QCLSPDSAGLVEDKLRLLACLYLSRPSYPKPLFQSLVSQLEQQGADVSAIHFLSR 252
C++ G EDKLR YLS S P Q+ L G + A++F+ R
Sbjct 416 ACINNKEQGTPEDKLRFFIIWYLSVDSVPASDLQAYEEALVNNGCTLEALNFVKR 470
> Hs7706371
Length=640
Score = 139 bits (350), Expect = 6e-33, Method: Compositional matrix adjust.
Identities = 84/228 (36%), Positives = 119/228 (52%), Gaps = 18/228 (7%)
Query 15 AEMFGSAATSLSKPLLILIDRDFDLLTMLQHTWLYGALIHDLLQLRLNRVT------VVT 68
+ G+ S +PLL+L+DR+ DL T L HTW Y AL+HD+L LNRV V
Sbjct 241 GDTLGAGQFSFQRPLLVLVDRNIDLATPLHHTWTYQALVHDVLDFHLNRVNLEESSGVEN 300
Query 69 PGEGGKP--STTKTFDLEKRDVFWREHAFSPFPVAAAAVSQMLTEYNAELAKVGQRPPDA 126
G +P K++DL D FW++H SPFP A +V Q L Y A+ +V +
Sbjct 301 SPAGARPKRKNKKSYDLTPVDKFWQKHKGSPFPEVAESVQQELESYRAQEDEVKRLKSIM 360
Query 127 FAQPE--------GDLSADILKAVDVLPDLADKKRSLDAHTTIATALVSAIKDRGLDRYF 178
+ E D +A + AV LP+L +KKR +D HT +ATA++ IK R LD YF
Sbjct 361 GLEGEDEGAISMLSDNTAKLTSAVSSLPELLEKKRLIDLHTNVATAVLEHIKARKLDVYF 420
Query 179 EIEQSLPQDRETVVTSAVQQCLSPDSAGLVEDKLRLLACLYLSRPSYP 226
E E+ + +T + ++ +S AG EDK+RL Y+S P
Sbjct 421 EYEEKIMS--KTTLDKSLLDIISDPDAGTPEDKMRLFLIYYISTQQAP 466
> Hs7706407
Length=638
Score = 137 bits (345), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 83/228 (36%), Positives = 118/228 (51%), Gaps = 18/228 (7%)
Query 15 AEMFGSAATSLSKPLLILIDRDFDLLTMLQHTWLYGALIHDLLQLRLNRVT------VVT 68
+ G+ S +PLL+L+DR+ DL T L HTW Y AL+HD+L LNRV V
Sbjct 240 GDTLGAGQFSFQRPLLVLVDRNIDLATPLHHTWTYQALVHDVLDFHLNRVNLEESSGVEN 299
Query 69 PGEGGKP--STTKTFDLEKRDVFWREHAFSPFPVAAAAVSQMLTEYNAELAKVGQRPPDA 126
G +P K++DL D FW++H SPFP A +V Q L Y A+ +V +
Sbjct 300 SPAGARPKRKNKKSYDLTPVDKFWQKHKGSPFPEVAESVQQELESYRAQEDEVKRLKSIM 359
Query 127 FAQPE--------GDLSADILKAVDVLPDLADKKRSLDAHTTIATALVSAIKDRGLDRYF 178
+ E D +A + AV LP+L +KKR +D HT +ATA++ IK R LD YF
Sbjct 360 GLEGEDEGAISMLSDNTAKLTSAVSSLPELLEKKRLIDLHTNVATAVLEHIKARKLDVYF 419
Query 179 EIEQSLPQDRETVVTSAVQQCLSPDSAGLVEDKLRLLACLYLSRPSYP 226
E E+ + +T + ++ +S AG ED +RL Y+S P
Sbjct 420 EYEEKIMS--KTTLDKSLLDIISDPDAGTPEDNMRLFLIYYISTQQAP 465
> 7295949
Length=641
Score = 135 bits (339), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 85/245 (34%), Positives = 128/245 (52%), Gaps = 15/245 (6%)
Query 19 GSAATSLSKPLLILIDRDFDLLTMLQHTWLYGALIHDLLQLRLNRVTV--VTPGEGGKPS 76
G S +P+L+L+DR+ DL T L HTW Y AL+HD+L L LN V V T KP
Sbjct 224 GGGVFSFQRPVLLLLDRNMDLATPLHHTWSYQALVHDVLDLGLNLVYVEDETARARKKP- 282
Query 77 TTKTFDLEKRDVFWREHAFSPFPVAAAAVSQMLTEYNAELAKVGQRPPDAFAQPEGDL-- 134
K DL++ D FW H SPFP A A+ + L Y ++ + + E D+
Sbjct 283 --KACDLDRNDRFWMTHKGSPFPTVAEAIQEELESYRNSEEEIKRLKTSMGIEGESDIAF 340
Query 135 ------SADILKAVDVLPDLADKKRSLDAHTTIATALVSAIKDRGLDRYFEIEQSLPQDR 188
+A + AV+ LP L +KKR +D HT IATA+++ IK R LD +FEIE+ + +
Sbjct 341 SLVNDTTARLTNAVNSLPQLMEKKRLIDMHTKIATAILNFIKARRLDSFFEIEEKV-MSK 399
Query 189 ETVVTSAVQQCLSPDSAGLVEDKLRLLACLYLSRPSYPKPLFQSLVSQLEQQGADVSAIH 248
+T+ + + G EDKLRL ++ P+ + L L+ G D++A+
Sbjct 400 QTLDRPLLDLLRDGE-FGQAEDKLRLYIIYFICAQQLPESEQERLKEALQAAGCDLTALA 458
Query 249 FLSRF 253
++ R+
Sbjct 459 YVQRW 463
> YDR189w
Length=666
Score = 125 bits (314), Expect = 9e-29, Method: Compositional matrix adjust.
Identities = 77/243 (31%), Positives = 123/243 (50%), Gaps = 17/243 (6%)
Query 24 SLSKPLLILIDRDFDLLTMLQHTWLYGALIHDLLQLRLNRVTV-VTPGEGG------KPS 76
SL + +LI++DR+ D +M H+W+Y ++ D+ +L N VT+ + E G KP
Sbjct 255 SLERGVLIILDRNIDFASMFSHSWIYQCMVFDIFKLSRNTVTIPLESKENGTDNTTAKPL 314
Query 77 TTKTFDLEKRDVFWREHAFSPFPVAAAAVSQMLTEYNAELAKVGQRP-----PDAFAQPE 131
TK +D+E D FW E++ PFP AA V L Y E A++ ++ D
Sbjct 315 ATKKYDIEPNDFFWMENSHLPFPEAAENVEAALNTYKEEAAEITRKTGVTNISDLDPNSN 374
Query 132 GDLSADILKAVDVLPDLADKKRSLDAHTTIATALVSAIKDRGLDRYFEIEQSLPQDRETV 191
D + I + V LP+L KK ++D H I AL+S ++ + LD +FE+EQ P +T
Sbjct 375 ND-TVQIQEVVKKLPELTAKKNTIDTHMNIFAALLSQLESKSLDTFFEVEQD-PGSTKT- 431
Query 192 VTSAVQQCLSPDSAGLVEDKLRLLACLYL-SRPSYPKPLFQSLVSQLEQQGADVSAIHFL 250
S L +EDKLR LYL S PK Q++ + ++ D++A+ ++
Sbjct 432 -RSRFLDILKDGKTNNLEDKLRSFIVLYLTSTTGLPKDFVQNVENYFKENDYDINALKYV 490
Query 251 SRF 253
+
Sbjct 491 YKL 493
> CE00993
Length=707
Score = 116 bits (291), Expect = 4e-26, Method: Compositional matrix adjust.
Identities = 80/244 (32%), Positives = 119/244 (48%), Gaps = 22/244 (9%)
Query 26 SKPLLILIDRDFDLLTMLQHTWLYGALIHDLLQLRLNRVTVVTPGEGGKPSTTKTFDLEK 85
S+PLL++ DR DL TML HTW Y AL+HD+L+L NRVT+ T G K K +D+
Sbjct 276 SRPLLVIGDRSADLATMLHHTWTYQALMHDVLELDQNRVTISTSSTGKK----KDYDMAT 331
Query 86 --RDVFWREHAFSPFPVAAAAVSQMLTEYNAE------------LAKVGQRPPDAFAQPE 131
D W H S FP A AV + L +Y ++ L+ + +A
Sbjct 332 GGSDKLWNNHKGSAFPTVAEAVQENLDDYRSKEDDIKKIKQALGLSGESEAADEALTNVL 391
Query 132 GDLSADILKAVDVLPDLADKKRSLDAHTTIATALVSAIKDRGLDRYFEIEQSLPQDRETV 191
D +A + V LPDL + KR +D HT IAT L+ IK+R LD +S V
Sbjct 392 ADTTAKLGSTVMSLPDLLESKRLIDLHTNIATTLLDVIKERKLDVSLSSSKSCCNTHRLV 451
Query 192 --VTSAVQQCLSPDSAGLVEDKLRLLACLYLSRPSYPKPLFQSLVSQLEQQGADVSAIHF 249
V Q LS + ED LR+L +L + + + + +++ L ++G + SA+
Sbjct 452 DLFYQPVTQFLS--NINHQEDVLRVLIIAFLCQETVNQNGYDQMMNLLRERGIEESALKH 509
Query 250 LSRF 253
+ +
Sbjct 510 VQKL 513
> At2g17980
Length=627
Score = 109 bits (272), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 72/242 (29%), Positives = 126/242 (52%), Gaps = 15/242 (6%)
Query 19 GSAATSLSKPLLILIDRDFDLLTMLQHTWLYGALIHDLLQLRLNRVTVVTPGEGGKPSTT 78
G +S +PLL + DR+F+L +QH + Y L+HD+L L+LN++ V GE G P
Sbjct 231 GGFMSSFQRPLLCIFDRNFELSVGIQHDFRYRPLVHDVLGLKLNQLKV--QGEKGPP--- 285
Query 79 KTFDLEKRDVFWREHAFSPFPVAAAAVSQMLTEYNAELAKVGQRP-PDAFAQPEG-DLSA 136
K+F+L+ D FW ++ FP A + L +Y ++ +V ++ + A+ +G DL
Sbjct 286 KSFELDSSDPFWSANSTLEFPDVAVEIETQLNKYKRDVEEVNKKTGGGSGAEFDGTDLIG 345
Query 137 DI-----LKAVDVLPDLADKKRSLDAHTTIATALVSAIKDRGLDRYFEIEQSLPQDRETV 191
+I + V LP+L ++K+ +D HT IATAL+ IK+R +D + + E + R +
Sbjct 346 NIHTEHLMNTVKSLPELTERKKVIDKHTNIATALLGQIKERSIDAFTKKESDMMM-RGGI 404
Query 192 VTSAVQQCLSPDSAGLVEDKLRLLACLYLSRPSYPKPLFQSLVSQLEQQGADVSAIHFLS 251
+ + L G DKLR +S + + +++ + L + AD SA ++
Sbjct 405 DRTELMAALK--GKGTKMDKLRFAIMYLISTETINQSEVEAVEAALNEAEADTSAFQYVK 462
Query 252 RF 253
+
Sbjct 463 KI 464
> ECU03g0900
Length=521
Score = 98.6 bits (244), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 64/199 (32%), Positives = 103/199 (51%), Gaps = 27/199 (13%)
Query 27 KPLLILIDRDFDLLTMLQHTWLYGALIHDLLQLRLNRVTVVTPGEGGKPSTTKTFDLEKR 86
+P+L+L+ R D++T +QH W Y AL++DL L N++T+ + K FDL+ +
Sbjct 203 RPVLVLVSRSHDVITPVQHVWSYSALMNDLFALESNKITL---------KSGKVFDLDPQ 253
Query 87 DVFWREHAFSPFPVAAAAVSQMLTEYNAELAKVGQRPPDAFAQPEGDLSADILKAVDVLP 146
D WR +A FPV V + L EY E+A R D E I +A+D P
Sbjct 254 DELWRRNANEYFPVVVERVEKELLEYKKEMA---LRSID-----EKTDKKVIQEALDKAP 305
Query 147 DLADKKRSLDAHTTIATALVSAIKDRGLDRYFEIEQSLPQDRETVVTSAVQQCLSPDSAG 206
+LA + S++AH +I + +V IK+R +D ++++E+ ++E + S G
Sbjct 306 ELAKRNESVNAHISICSEMVEMIKERAIDDFYKVEKGGHTNQELIELS---------EKG 356
Query 207 LVEDKLRLLACLYLSRPSY 225
ED LR LA L L+ +
Sbjct 357 SDEDVLR-LAILLLNTKDF 374
> At1g02010
Length=426
Score = 68.9 bits (167), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 43/156 (27%), Positives = 70/156 (44%), Gaps = 6/156 (3%)
Query 30 LILIDRDFDLLTMLQHTWLYGALIHDLLQLRLNRVTVVTPGEGGKPSTTKTFDLEKRDVF 89
L+++DR D + + H W Y A+ HDLL + N+ + P + G P K LE D
Sbjct 27 LLIVDRSVDQIAPIIHEWTYDAMCHDLLDMEGNKHVIEVPSKTGGPPEKKEIVLEDHDPV 86
Query 90 WREHAFSPFPVAAAAVSQMLTEYNAELAKVGQRPPDAFAQPEGDLSA-DILKAVDVLPDL 148
W E + A+ + + +T + ++ R D +LS D+ K V LP
Sbjct 87 WLELRHTHIADASERLHEKMTNFASKNKAAQMRSRDG-----SELSTRDLQKIVQALPQY 141
Query 149 ADKKRSLDAHTTIATALVSAIKDRGLDRYFEIEQSL 184
++ L H +A + I+D GL ++EQ L
Sbjct 142 GEQVDKLSTHVELAGKINRIIRDTGLRDLGQLEQDL 177
> At1g12360
Length=637
Score = 66.2 bits (160), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 61/230 (26%), Positives = 98/230 (42%), Gaps = 20/230 (8%)
Query 30 LILIDRDFDLLTMLQHTWLYGALIHDLLQLRLNRVTVVTPGEGGKPSTTKTFDLEKRDVF 89
L+++DR D + + H W Y A+ HDLL + N+ V P + G K LE+ D
Sbjct 226 LLILDRSIDQIAPVIHEWTYDAMCHDLLNMEGNKYVHVIPSKSGGQPEKKDVLLEEHDPI 285
Query 90 WREHAFSPFPVAAAAVSQMLTEY---NAELAKVGQRPPDAFAQPEGDLSADILKAVDVLP 146
W E + A+ + +T + N G+R + + D+ K V LP
Sbjct 286 WLELRHAHIADASERLHDKMTNFLSKNKAAQLQGKRDGAELS------TRDLQKMVQALP 339
Query 147 DLADKKRSLDAHTTIATALVSAIKDRGLDRYFEIEQSLPQDRETVVTSA----VQQCLSP 202
+++ L H IA L I+++GL ++EQ L V A V + LS
Sbjct 340 QYSEQIDKLSLHVEIARKLNDLIREQGLRELGQLEQDL------VFGDAGMKDVIKYLST 393
Query 203 DSAGLVEDKLRLLACLYLSRP-SYPKPLFQSLVSQLEQQGADVSAIHFLS 251
E KLRLL L P + Q+L+ + D++A++ +S
Sbjct 394 QEEASREGKLRLLMILATIYPEKFEGEKGQNLMKLAKLSSDDMTAVNNMS 443
> At4g12120
Length=635
Score = 63.9 bits (154), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 54/192 (28%), Positives = 88/192 (45%), Gaps = 16/192 (8%)
Query 30 LILIDRDFDLLTMLQHTWLYGALIHDLLQLRLNRVTVVTPGEGGKPSTTKTFDLEKRDVF 89
L+++DR D + L H W Y A+ HDLL + N+ T P + G K L++ D
Sbjct 233 LLILDRSIDQIAPLIHEWTYDAMCHDLLNMEGNKYTHEVPSKTGDKPEKKEVLLDEEDSI 292
Query 90 WREHAFSPFPVAAAAVSQMLTEY--NAELAKVGQRPPDAFAQPEGDLSA-DILKAVDVLP 146
W E + A+ + + +T + + A++ D GDLS+ D+ K V LP
Sbjct 293 WVELRDAHIADASERLHEKMTNFVSKNKAAQLKHSSKDF-----GDLSSKDLQKMVHALP 347
Query 147 DLADKKRSLDAHTTIATALVSAIKDRGLDRYFEIEQSL---PQDRETVVTSAVQQCLSPD 203
+++ L H IA + I ++GL ++EQ L R+ V+ + LS +
Sbjct 348 QYSEQIDKLSLHVEIARTINRTIMEQGLRDLGQLEQDLVFGDAGRKDVI-----KFLSTN 402
Query 204 SAGLVEDKLRLL 215
E KLRL+
Sbjct 403 HIISHESKLRLI 414
> At1g77140
Length=550
Score = 62.4 bits (150), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 55/217 (25%), Positives = 99/217 (45%), Gaps = 18/217 (8%)
Query 26 SKPLLILIDRDFDLLTMLQHTWLYGALIHDLLQLRLNRVTVVTPGEGGKPSTTKTFDLEK 85
S PLL++IDR D +T L + W Y A++H+L+ L+ N+V + + G K + +
Sbjct 218 SSPLLLVIDRRDDPVTPLLNQWTYQAMVHELIGLQDNKVDLKSIGSLPKDQQVEVVLSSE 277
Query 86 RDVFWREHAFSPFPVAAAAVSQMLTEYNAELAKVGQRPPDAFAQPEGDLSADILKAVDVL 145
+D F++ + + F + +M+ ++ ++AK Q D+ + VD
Sbjct 278 QDAFFKSNMYENFGDIGMNIKRMVDDFQ-QVAKSNQNIQTV---------EDMARFVDNY 327
Query 146 PDLADKKRSLDAHTTIATALVSAIKDRGLDRYFEIEQSLP-QDRETVVTSAVQQCLSPDS 204
P+ + ++ H T+ T + ++ R L + EQ L + AV L+ +S
Sbjct 328 PEYKKMQGNVSKHVTLVTEMSKLVEARKLMTVSQTEQDLACNGGQGAAYEAVTDLLNNES 387
Query 205 AGLVEDKLRLLACLYLSRPSYPKP-----LFQSLVSQ 236
+ D+LRL+ LY R P LF L S+
Sbjct 388 VSDI-DRLRLV-MLYALRYEKENPVQLMQLFNKLASR 422
> 7299206
Length=574
Score = 53.9 bits (128), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 46/203 (22%), Positives = 86/203 (42%), Gaps = 19/203 (9%)
Query 18 FGSAATSLSKPLLILIDRDFDLLTMLQHTWLYGALIHDLLQLRLNRVTVVTPGEGGKPST 77
F S + PLL+++DR D +T L H W Y A++H+LL ++ NR+ + P+
Sbjct 205 FRSNMDGAAPPLLLVLDRRDDPVTPLLHQWTYQAMVHELLHIKNNRLDL-----SNCPNV 259
Query 78 TKTFD----LEKRDVFWREHAFSPFPVAAAAVSQMLTEYNAELAKVGQRPPDAFAQPEGD 133
K F +D F+ + ++ + + + Q++ E+ QR + + E
Sbjct 260 PKDFKELVLSGDQDDFYGNNMYANYGEIGSTIKQLMEEF--------QRKANDHKKVES- 310
Query 134 LSADILKAVDVLPDLADKKRSLDAHTTIATALVSAIKDRGLDRYFEIEQSLPQDRETVVT 193
AD+ ++ P ++ H + L + R L E+EQ + E
Sbjct 311 -IADMKNFIESYPQFKKMSGTVQKHLCVIGELSALSNKRNLFEVSELEQEIACKAEHSAQ 369
Query 194 SAVQQCLSPDSAGLVEDKLRLLA 216
+ L D ++D L+L+A
Sbjct 370 LQRIKKLIADERVSIDDALKLVA 392
> Hs18105063
Length=570
Score = 46.2 bits (108), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 52/232 (22%), Positives = 98/232 (42%), Gaps = 26/232 (11%)
Query 1 RLQELLEALPPKAFAEMFGSAATSLSKPLLILIDRDFDLLTMLQHTWLYGALIHDLLQLR 60
RL E ++ + K + E+F T + PLL+++DR D +T L + W Y A++H+LL +
Sbjct 186 RLAECVKQVITKEY-ELFEFRRTEVP-PLLLILDRCDDAITPLLNQWTYQAMVHELLGIN 243
Query 61 LNRVTVVTPGEGGKPSTTKTFDLE------KRDVFWREHAFSPFPVAAAAVSQMLTEYNA 114
NR+ + P +K DL + D F+ + + F + + ++ ++
Sbjct 244 NNRIDL-----SRVPGISK--DLREVVLSAENDEFYANNMYLNFAEIGSNIKNLMEDFQK 296
Query 115 ELAKVGQRPPDAFAQPEGDLSADILKAVDVLPDLADKKRSLDAHTTIATALVSAIKDRGL 174
+ K Q+ AD+ V+ P ++ H T+ L + +R L
Sbjct 297 KKPKEQQKLESI---------ADMKAFVENYPQFKKMSGTVSKHVTVVGELSRLVSERNL 347
Query 175 DRYFEIEQSLP--QDRETVVTSAVQQCLSPDSAGLVEDKLRLLACLYLSRPS 224
E+EQ L D + + + + +P +L +L L+ R S
Sbjct 348 LEVSEVEQELACQNDHSSALQNIKRLLQNPKVTEFDAARLVMLYALHYERHS 399
> SPAC2G11.03c
Length=558
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 34/157 (21%), Positives = 66/157 (42%), Gaps = 10/157 (6%)
Query 28 PLLILIDRDFDLLTMLQHTWLYGALIHDLLQLRLNRVTVVTPGEGGKPSTTKTFDLEKRD 87
P+L+L+DR D +T L W Y A++H+L + RV+ + ST + D
Sbjct 211 PILLLLDRKNDPITPLLTQWTYQAMVHELFGIDNGRVSFSNSTSDNEKSTEIVLN-PTLD 269
Query 88 VFWREHAFSPFPVAAAAVSQMLTEYNAELAKVGQRPPDAFAQPEGDLSADILKAVDVLPD 147
F++E F F + ++ + K E + AD+ + ++ P+
Sbjct 270 PFYKETRFDNFGDLGVKIKDYVSHLQTKSTKKAS---------EIESIADMKQFLEAYPE 320
Query 148 LADKKRSLDAHTTIATALVSAIKDRGLDRYFEIEQSL 184
++ H ++ + + ++ L E+EQSL
Sbjct 321 YRRLSGNVSKHVSLVSEISQVVQRENLLEVGEVEQSL 357
> CE04214
Length=547
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 49/223 (21%), Positives = 90/223 (40%), Gaps = 21/223 (9%)
Query 19 GSAATSLSKPLLILIDRDFDLLTMLQHTWLYGALIHDLLQLRLNRVTVVTPGEGGKPSTT 78
G S + L++I+R D +T L + W Y A+IH++L L NR T T
Sbjct 202 GLFENSRADTTLLIIERSQDAVTPLLNQWTYEAMIHEMLTLTNNRCTC----------TD 251
Query 79 KTFDL-EKRDVFWREHAFSPFPVAAAAVSQMLTEYNAELAKVGQRPPDAFAQPEGDLSAD 137
+ L E D F+ + + F + +++E+ E + + D
Sbjct 252 QNIVLSELHDEFFARNITANFGEIGQNIKTLISEF-QEKKHINKNLESI---------QD 301
Query 138 ILKAVDVLPDLADKKRSLDAHTTIATALVSAIKDRGLDRYFEIEQSLPQDRETVVTSAVQ 197
+ K V+ P ++ H ++ L + ++ L E+EQ++ D + +
Sbjct 302 MKKFVEDYPQFKKISGTVSKHVSLVGELSNLVQKHNLLGVSEVEQAIVSDGDHGKCINLV 361
Query 198 QCLSPDSAGLVEDKLRLLACLYLSRPSYPKPLFQSLVSQLEQQ 240
+ L ++ D +RL+ L + P SL+SQL Q
Sbjct 362 RGLLKNTKTREVDIIRLVLLYALRFENAPGNELNSLISQLRPQ 404
> YGL095c
Length=577
Score = 42.7 bits (99), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 54/229 (23%), Positives = 95/229 (41%), Gaps = 22/229 (9%)
Query 26 SKPLLILIDRDFDLLTMLQHTWLYGALIHDLLQLRLNRVTVVTPGEGGKPSTTKTFDLEK 85
S P+L+++DR+ D +T L W Y ++I++ + ++ N V + K T K
Sbjct 223 STPVLLILDRNTDPITPLLQPWTYQSMINEYIGIKRNIVDLSKVPRIDKDLEKVTLS-SK 281
Query 86 RDVFWREHAFSPFPVAAAAVSQMLTEYNAELAKVGQRPPDAFAQPEGDLSA--DILKAVD 143
+D F+R+ + F V Q +T Y + Q +++ DI ++
Sbjct 282 QDAFFRDTMYLNFGELGDKVKQYVTTYKDK------------TQTNSQINSIEDIKNFIE 329
Query 144 VLPDLADKKRSLDAHTTIATALVSAIKDRGLDRYFEIEQSLP-QDRETVVTSAVQQCLSP 202
P+ ++ H I L +K + + EIEQ+L D S + + L
Sbjct 330 KYPEFRKLSGNVAKHMAIVGELDRQLKIKNIWEISEIEQNLSAHDANEEDFSDLIKLLQN 389
Query 203 DSAGLVEDKLRL-LACLYLSRPSYPKPLFQSLVSQLEQQ--GADVSAIH 248
++ V+ +L LAC+Y + LV L QQ DV+ H
Sbjct 390 EA---VDKYYKLKLACIYSLNNQTSSDKIRQLVEILSQQLPPEDVNFFH 435
> CE24927
Length=591
Score = 40.0 bits (92), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 38/155 (24%), Positives = 66/155 (42%), Gaps = 11/155 (7%)
Query 30 LILIDRDFDLLTMLQHTWLYGALIHDLLQLRLNRVTVVTPGEGGKPSTTKTFDLEKRDVF 89
LI+IDR +D +T L H A+ +DLL + N V G G + K L++ D
Sbjct 228 LIIIDRGYDAITPLLHELTLQAMCYDLLGIE-NDVYKYETG-GSDENLEKEVLLDENDDL 285
Query 90 WREHAFSPFPVAAAAVSQMLTEYNAELAKVGQRPPDAFAQPEGDLSADILKAVDVLPDLA 149
W E V + V++ L +++ G + DLS I + +P
Sbjct 286 WVEMRHKHIAVVSQEVTKNLKKFSESKGNKGTMDSKSIK----DLSMLIKR----MPQHK 337
Query 150 DKKRSLDAHTTIATALVSAIKDRGLDRYFEIEQSL 184
+ H ++A + + +G+D+ ++EQ L
Sbjct 338 KELNKFSTHISLAEECMKQYQ-QGVDKLCKVEQDL 371
> 7292440
Length=597
Score = 35.0 bits (79), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 26/83 (31%), Positives = 42/83 (50%), Gaps = 4/83 (4%)
Query 30 LILIDRDFDLLTMLQHTWLYGALIHDLLQLRLNRVTVVTPGEGGKPSTTKTFDLEKRDVF 89
L+++DR FD ++ L H A+ +DLL + +N V TPG +P K L++ D
Sbjct 241 LLILDRGFDCVSPLLHELTLQAMAYDLLPI-VNDVYRYTPGP-NQPD--KEVLLDENDDL 296
Query 90 WREHAFSPFPVAAAAVSQMLTEY 112
W E V + V+Q L ++
Sbjct 297 WVELRHEHIAVVSTQVTQNLKKF 319
> Hs18105056
Length=596
Score = 34.7 bits (78), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 43/199 (21%), Positives = 78/199 (39%), Gaps = 23/199 (11%)
Query 30 LILIDRDFDLLTMLQHTWLYGALIHDLLQLRLNRVTV-------VTPGEGGK--PSTTKT 80
L+L+DR+ DLLT L Y LI ++ ++ + V + G+GGK P+ K
Sbjct 227 LLLLDRNVDLLTPLATQLTYEGLIDEIYGIQNSYVKLPPEKFAPKKQGDGGKDLPTEAKK 286
Query 81 FDLEKRDVFWREHAFSPFPVAAAAVSQMLTEYNAELAKVGQRPPDAFAQPEGDLSADILK 140
L + + E F + +S+ +A + + +I +
Sbjct 287 LQLNSAEELYAEIRDKNFNAVGSVLSKKAKIISAAFEE----------RHNAKTVGEIKQ 336
Query 141 AVDVLPDLADKKRSLDAHTTIATALVSAIKDRGLDRYFEIEQSLPQDRET-VVTSAVQQC 199
V LP + + SL HT+IA + +EQ +T V + ++ C
Sbjct 337 FVSQLPHMQAARGSLANHTSIAELIKDVTTSEDFFDKLTVEQEFMSGIDTDKVNNYIEDC 396
Query 200 LSPDSAGLVEDKLRLLACL 218
++ + + K+ L CL
Sbjct 397 IAQKHSLI---KVLRLVCL 412
> 7293600
Length=617
Score = 34.3 bits (77), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 41/160 (25%), Positives = 67/160 (41%), Gaps = 23/160 (14%)
Query 30 LILIDRDFDLLTMLQHTWLYGALIHDLLQLRLNRVTVVT---PGEGGKP----------- 75
LIL+DR DLL+ L Y LI + +R N++T+ P +G P
Sbjct 228 LILLDRSIDLLSPLATQLTYEGLIDEFYGIRQNKLTLPAENFPSDGALPGGGGSGPRVEE 287
Query 76 STTKTFDLEKRDVFWREHAFSPFPVAAAAVSQMLTEYNAELAKVGQRPPDAFAQPEGDLS 135
S + D EK+ + S + A ++ E LA+ + D S
Sbjct 288 SQSLLGDTEKKTILLH----SGEQLYAELRNKHFNEVTKLLARKAREIHVQMHATSQDKS 343
Query 136 ADILKAV--DVLPDLADKKRSLDAHTTIATAL---VSAIK 170
+K+ ++LP L +K++ HT IA L V+A++
Sbjct 344 VQEIKSFVENLLPQLMAQKKATSEHTAIAGLLHEQVNAVR 383
> Hs6005886
Length=592
Score = 32.0 bits (71), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 21/82 (25%), Positives = 37/82 (45%), Gaps = 6/82 (7%)
Query 30 LILIDRDFDLLTMLQHTWLYGALIHDLLQLRLNRVTVVTPGEGGKPSTTKTFDLEKRDVF 89
L++IDR FD ++ + H + A+ +DLL + + T G+G K LE+ D
Sbjct 235 LLIIDRGFDPVSTVLHELTFQAMAYDLLPIENDTYKYKTDGKG------KEAILEEEDDL 288
Query 90 WREHAFSPFPVAAAAVSQMLTE 111
W V + +++ E
Sbjct 289 WVRIRHRHIAVVLEEIPKLMKE 310
> SPCC584.05
Length=693
Score = 31.6 bits (70), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 17/57 (29%), Positives = 25/57 (43%), Gaps = 0/57 (0%)
Query 26 SKPLLILIDRDFDLLTMLQHTWLYGALIHDLLQLRLNRVTVVTPGEGGKPSTTKTFD 82
SK +++DR D H + Y A+IHDLL ++ + G G T D
Sbjct 225 SKSTCLIVDRSLDTAAPFLHEFTYQAMIHDLLPIKNEQYPYEILGPQGTEKRTGKLD 281
> ECU06g1460
Length=490
Score = 31.2 bits (69), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 13/26 (50%), Positives = 19/26 (73%), Gaps = 0/26 (0%)
Query 30 LILIDRDFDLLTMLQHTWLYGALIHD 55
LI++DR FDL T L + W Y +L+H+
Sbjct 200 LIMLDRAFDLYTPLLYEWRYQSLLHE 225
> Hs11342680
Length=376
Score = 30.0 bits (66), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 27/84 (32%), Positives = 37/84 (44%), Gaps = 7/84 (8%)
Query 125 DAFAQPEGDLSADIL-KAVDVLPDLADKKRSLDAHTTIATALVSAIKDRGLDRYFEIEQS 183
+ FA P + DI + V L +K +D HT+ +V IK+R Y I
Sbjct 172 EGFAMPHSIMRVDIAGRDVSRYLRLLLRKEGVDFHTSAEFEVVRTIKERAC--YLSIN-- 227
Query 184 LPQDRETVVTSAVQQCLSPDSAGL 207
PQ E + T VQ L PD + L
Sbjct 228 -PQKDEALETEKVQYTL-PDGSTL 249
> CE13120
Length=1521
Score = 29.3 bits (64), Expect = 8.3, Method: Composition-based stats.
Identities = 16/50 (32%), Positives = 23/50 (46%), Gaps = 0/50 (0%)
Query 74 KPSTTKTFDLEKRDVFWREHAFSPFPVAAAAVSQMLTEYNAELAKVGQRP 123
K S K DL +RD +W EH + + S+ TE N + +V P
Sbjct 1252 KSSDLKEDDLSQRDQYWDEHQYQADLELMKSESKKKTEANTRVKRVIAHP 1301
Lambda K H
0.320 0.135 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5223052002
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40