bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1066_orf1
Length=159
Score E
Sequences producing significant alignments: (Bits) Value
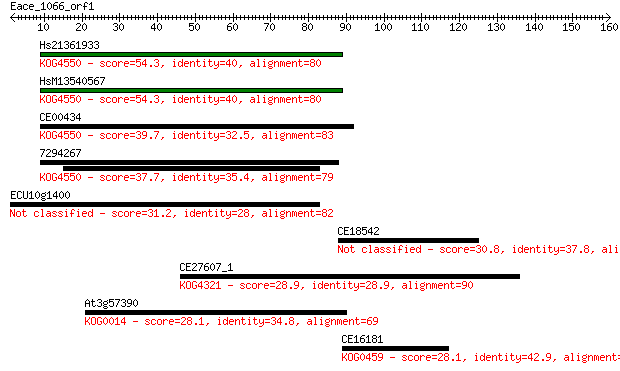
Hs21361933 54.3 1e-07
HsM13540567 54.3 1e-07
CE00434 39.7 0.002
7294267 37.7 0.010
ECU10g1400 31.2 0.80
CE18542 30.8 1.0
CE27607_1 28.9 4.6
At3g57390 28.1 7.6
CE16181 28.1 8.2
> Hs21361933
Length=612
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 32/82 (39%), Positives = 44/82 (53%), Gaps = 6/82 (7%)
Query 9 PHSSAVADIDGDCRADLLLMVQSPDSSSVSLEVWLNRYDG--AAATWERQQQNIALPPGS 66
PHS A D+ D ADL L + +S+ E+W N DG + +T + QN+ +
Sbjct 204 PHSHAFIDLTEDFTADLFLTTLNATTSTFQFEIWEN-LDGNFSVSTILEKPQNMMV---V 259
Query 67 GQILLADFDADGTLDILVPSCE 88
GQ ADFD DG +D L+P CE
Sbjct 260 GQSAFADFDGDGHMDHLLPGCE 281
> HsM13540567
Length=609
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 32/82 (39%), Positives = 44/82 (53%), Gaps = 6/82 (7%)
Query 9 PHSSAVADIDGDCRADLLLMVQSPDSSSVSLEVWLNRYDG--AAATWERQQQNIALPPGS 66
PHS A D+ D ADL L + +S+ E+W N DG + +T + QN+ +
Sbjct 201 PHSHAFIDLTEDFTADLFLTTLNATTSTFQFEIWEN-LDGNFSVSTILEKPQNMMV---V 256
Query 67 GQILLADFDADGTLDILVPSCE 88
GQ ADFD DG +D L+P CE
Sbjct 257 GQSAFADFDGDGHMDHLLPGCE 278
> CE00434
Length=599
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 27/87 (31%), Positives = 42/87 (48%), Gaps = 11/87 (12%)
Query 9 PHSSAVADIDGDCRADLLLMVQSPDSSSVSLEVWLNRYDGAAATWERQQQNIALPPGS-- 66
PH D++ D AD++ M + D S + + VW +W+ + L P
Sbjct 202 PH--LFVDLNSDLIADIVFMTKESDGS-LFMSVW----QKTKISWQFRDWVPKLTPAQYP 254
Query 67 --GQILLADFDADGTLDILVPSCEKNE 91
G ++ D D+DG LDILVP C ++E
Sbjct 255 FVGAPVVMDVDSDGELDILVPICREDE 281
> 7294267
Length=596
Score = 37.7 bits (86), Expect = 0.010, Method: Composition-based stats.
Identities = 28/86 (32%), Positives = 41/86 (47%), Gaps = 14/86 (16%)
Query 9 PHSSAVADIDGDCRADLLLMVQSPDSSSVSLEVW--LNRYDGAAATWERQQQNIALPPGS 66
P+++A D++GD ADL L +S S E+W +N D + Q Q P G
Sbjct 191 PNANAYLDLNGDFLADLFLQTKS------SYEIWHGINERD-KQENFSYQYQIKFQPIGG 243
Query 67 -----GQILLADFDADGTLDILVPSC 87
GQ + DF+ G +I+VP C
Sbjct 244 NDYYIGQAVFMDFELKGVQNIVVPFC 269
Score = 29.6 bits (65), Expect = 2.8, Method: Composition-based stats.
Identities = 18/69 (26%), Positives = 33/69 (47%), Gaps = 8/69 (11%)
Query 15 ADIDGDCRADLLLMVQSPDSSSVSLEV-WLNRYDGAAATWERQQQNIALPPGSGQILLAD 73
D DGD D+L+ ++ D + + + W N T E + N +L G+++ D
Sbjct 91 GDFDGDALMDVLVNLKGKDEETYEVHINWGN-------TTELKCSNKSLFTSKGEVMALD 143
Query 74 FDADGTLDI 82
F+ D +D+
Sbjct 144 FNRDMIIDL 152
> ECU10g1400
Length=561
Score = 31.2 bits (69), Expect = 0.80, Method: Composition-based stats.
Identities = 23/82 (28%), Positives = 39/82 (47%), Gaps = 6/82 (7%)
Query 1 EEEALITNPHSSAVADIDGDCRADLLLMVQSPDSSSVSLEVWLNRYDGAAATWERQQQNI 60
+E + H+SA ++DG +A L L+V + + L V NR +E +
Sbjct 152 DEYGKLHRDHTSAFVELDGSMKACLCLVVDDGNGGKL-LRVLSNRNGEFVNYFE-----M 205
Query 61 ALPPGSGQILLADFDADGTLDI 82
+LP G I+ DF+ +G D+
Sbjct 206 SLPNEIGPIVFGDFNGNGMNDM 227
> CE18542
Length=2962
Score = 30.8 bits (68), Expect = 1.0, Method: Composition-based stats.
Identities = 14/37 (37%), Positives = 20/37 (54%), Gaps = 3/37 (8%)
Query 88 EKNEEGVCIKNDALILMPNTQPPLCGSFWWTSRTVEK 124
+KN + C++N L MP + P S W RT+EK
Sbjct 1879 KKNAQSGCVENKGLSRMPKFEEPFVDSVW---RTIEK 1912
> CE27607_1
Length=426
Score = 28.9 bits (63), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 26/95 (27%), Positives = 38/95 (40%), Gaps = 8/95 (8%)
Query 46 YDGAAATWERQQ-QNIALPPGSGQILLADFDADGTLDILVP----SCEKNEEGVCIKNDA 100
+DG W + + I P G L + LD+ KN C+ +
Sbjct 102 WDGVGNVWHGYELRGIENVPDEGSALFIYYHGCLPLDVYYLISKLVIHKNRSLHCVGDKF 161
Query 101 LILMPNTQPPLCGSFWWTSRTVEKCRRPFELCDAN 135
+ +P +P LC F TS TVE+C EL + N
Sbjct 162 IFKIPGWRP-LCKLFSITSGTVEECTE--ELKEGN 193
> At3g57390
Length=256
Score = 28.1 bits (61), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 37/79 (46%), Gaps = 10/79 (12%)
Query 21 CRADLLLMVQSP-----DSSSVSLEVWLNRYDGAAATWERQQQN-----IALPPGSGQIL 70
C A++ L++ S D SSV +E L+RY A+ E +QQ I G+ +L
Sbjct 39 CDAEVALIIFSSTGKIYDFSSVCMEQILSRYGYTTASTEHKQQREHQLLICASHGNEAVL 98
Query 71 LADFDADGTLDILVPSCEK 89
D G L+ L + E+
Sbjct 99 RNDDSMKGELERLQLAIER 117
> CE16181
Length=573
Score = 28.1 bits (61), Expect = 8.2, Method: Composition-based stats.
Identities = 12/28 (42%), Positives = 14/28 (50%), Gaps = 0/28 (0%)
Query 89 KNEEGVCIKNDALILMPNTQPPLCGSFW 116
K E G K D L++MPN QP W
Sbjct 358 KMESGCVQKGDTLVVMPNKQPVQVLQIW 385
Lambda K H
0.315 0.132 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2136300674
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40