bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1086_orf2
Length=345
Score E
Sequences producing significant alignments: (Bits) Value
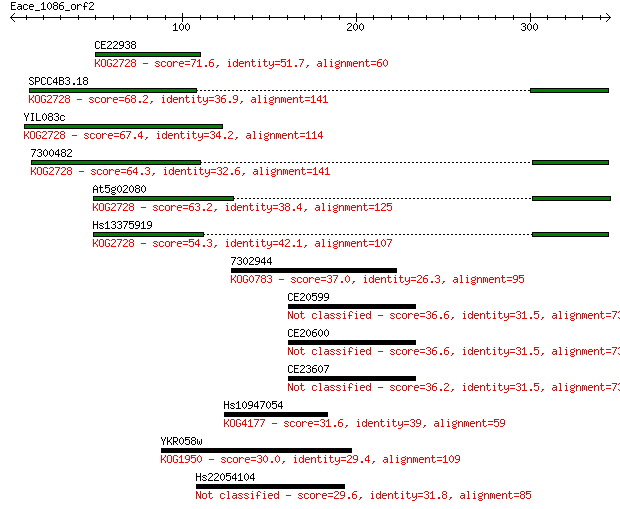
CE22938 71.6 3e-12
SPCC4B3.18 68.2 3e-11
YIL083c 67.4 4e-11
7300482 64.3 4e-10
At5g02080 63.2 8e-10
Hs13375919 54.3 4e-07
7302944 37.0 0.055
CE20599 36.6 0.071
CE20600 36.6 0.072
CE23607 36.2 0.097
Hs10947054 31.6 2.3
YKR058w 30.0 6.8
Hs22054104 29.6 8.7
> CE22938
Length=277
Score = 71.6 bits (174), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 31/60 (51%), Positives = 40/60 (66%), Gaps = 0/60 (0%)
Query 50 LAFITAGGTDVPLEREAVRYITNTSSGARGAALCEQLLRLGYYVVYLTSVRAMKPFVRHL 109
+ FIT+GGT VPLE+ VR+I N S G RGAA E LR GY V+++ ++KPF RH
Sbjct 19 IVFITSGGTQVPLEKNTVRFIDNFSMGTRGAASAEYFLRAGYAVIFMHREESLKPFSRHF 78
> SPCC4B3.18
Length=316
Score = 68.2 bits (165), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 34/96 (35%), Positives = 52/96 (54%), Gaps = 5/96 (5%)
Query 12 ADSFFAAEPPPPNVATAGLRVRQFVRPLLENEELKEAKLAFITAGGTDVPLEREAVRYIT 71
++ FF P P ++ + F+ N + +AF+T+GGT VPLE+ VR+I
Sbjct 7 SNDFFEENPCPNSLGDVSSLIEDFI-----NLQSNSQNIAFVTSGGTLVPLEQNTVRFID 61
Query 72 NTSSGARGAALCEQLLRLGYYVVYLTSVRAMKPFVR 107
N S+G RGAA E + GY V++L ++ PF R
Sbjct 62 NFSAGNRGAASAEYFCKSGYSVIFLYRNYSLMPFTR 97
Score = 40.4 bits (93), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 18/45 (40%), Positives = 29/45 (64%), Gaps = 0/45 (0%)
Query 300 RLLCITYRTLVDYAFIVRAVAAGAAPLRERLMFCSAAAVADFYLP 344
RL+ I Y TL +Y + ++A+A + R +F AAAV+DF++P
Sbjct 143 RLIMIPYTTLTEYLWYLKAIAERLQIIASRALFYLAAAVSDFHIP 187
> YIL083c
Length=365
Score = 67.4 bits (163), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 39/115 (33%), Positives = 60/115 (52%), Gaps = 6/115 (5%)
Query 9 TSSADSFFAAEPPPPNVATAGLRVRQFVRPLLENEELKEAKLAFITAGGTDVPLEREAVR 68
T+ + +F P P + ++F+ LK K+ IT+GGT VPLE VR
Sbjct 37 TTEEEQYFKTNPKPAYIDELIKDAKEFIDL---QYSLKRNKIVLITSGGTTVPLENNTVR 93
Query 69 YITNTSSGARGAALCEQLLRLGYYVVYLTSVRAMKPFVRHLLPTHPMPHM-LDFI 122
+I N S+G RGA+ EQ L GY V++L ++ P+ R +H + + LD+I
Sbjct 94 FIDNFSAGTRGASSAEQFLANGYSVIFLHREFSLTPYNRSF--SHSINTLFLDYI 146
> 7300482
Length=313
Score = 64.3 bits (155), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 29/97 (29%), Positives = 50/97 (51%), Gaps = 4/97 (4%)
Query 13 DSFFAAEPPPPNVATAGLRVRQFVRPLLENEELKEAKLAFITAGGTDVPLEREAVRYITN 72
+ F+ PP + R ++ E + ++ +T+GGT VPLE VR++ N
Sbjct 5 EDFYNTHLPPADFEDN----RSLLKEFCERHNKLQNRIVLVTSGGTTVPLEHNTVRFVDN 60
Query 73 TSSGARGAALCEQLLRLGYYVVYLTSVRAMKPFVRHL 109
S+G RG+A E L Y V+++ ++++PF RH
Sbjct 61 FSAGTRGSASAEYFLDHDYAVIFMHRHKSLEPFTRHF 97
Score = 39.7 bits (91), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 28/44 (63%), Gaps = 0/44 (0%)
Query 301 LLCITYRTLVDYAFIVRAVAAGAAPLRERLMFCSAAAVADFYLP 344
+L + + ++VDY +++RA A ER + AAAV+DFY+P
Sbjct 143 ILYVNFTSVVDYMWLLRAACECLAAFEERAVLYLAAAVSDFYIP 186
> At5g02080
Length=270
Score = 63.2 bits (152), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 32/80 (40%), Positives = 45/80 (56%), Gaps = 1/80 (1%)
Query 49 KLAFITAGGTDVPLEREAVRYITNTSSGARGAALCEQLLRLGYYVVYLTSVRAMKPFVRH 108
++ +T+GGT VPLE+ VRYI N SSG RGAA E ++ GY V++L +P+ R
Sbjct 23 RIVCVTSGGTTVPLEQRCVRYIDNFSSGNRGAASTENFVKAGYAVIFLYRRGTCQPYCRS 82
Query 109 LLPTHPMPHMLDFISLCTVQ 128
LP P +F +Q
Sbjct 83 -LPDDPFLECFEFSDKTNIQ 101
Score = 31.6 bits (70), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 16/45 (35%), Positives = 27/45 (60%), Gaps = 0/45 (0%)
Query 301 LLCITYRTLVDYAFIVRAVAAGAAPLRERLMFCSAAAVADFYLPY 345
LL + + T+ +Y ++R +A + MF AAAV+DFY+P+
Sbjct 104 LLKLPFSTIYEYLQMLRLIAEALKDVGPCSMFYLAAAVSDFYVPW 148
> Hs13375919
Length=284
Score = 54.3 bits (129), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 26/63 (41%), Positives = 36/63 (57%), Gaps = 0/63 (0%)
Query 49 KLAFITAGGTDVPLEREAVRYITNTSSGARGAALCEQLLRLGYYVVYLTSVRAMKPFVRH 108
++ +T+GGT VPLE VR++ N SSG RGA E L GY V++L R+ P+
Sbjct 36 RVVLVTSGGTKVPLEARPVRFLDNFSSGRRGATSAEAFLAAGYGVLFLYRARSAFPYAHR 95
Query 109 LLP 111
P
Sbjct 96 FPP 98
Score = 38.9 bits (89), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 19/44 (43%), Positives = 26/44 (59%), Gaps = 0/44 (0%)
Query 301 LLCITYRTLVDYAFIVRAVAAGAAPLRERLMFCSAAAVADFYLP 344
L + + TL DY +++A A PL MF AAAV+DFY+P
Sbjct 144 FLAVEFTTLADYLHLLQAAAQALNPLGPSAMFYLAAAVSDFYVP 187
> 7302944
Length=1187
Score = 37.0 bits (84), Expect = 0.055, Method: Composition-based stats.
Identities = 25/97 (25%), Positives = 42/97 (43%), Gaps = 4/97 (4%)
Query 128 QHQQKEEGQEEVEEEEQQPQPIVEEPSSDTDASECEEGVSSPSPPPSSRHLACQTFSAVY 187
+H+QK+ +++ Q Q + SE +G PS P S + +S V
Sbjct 903 KHKQKDLRKQDARH--QYEQQAISSMMRSLSISESTQGTEVPSSPQDSARSEDKNWSRVV 960
Query 188 EAEEEPD--AQVVTSVGSVLKSSDQPTDENAPAEAEP 222
+ +++ A+ V + LK D PT E P E +P
Sbjct 961 DKKDQKRKLAETALKVNNTLKHEDPPTQELVPIERKP 997
> CE20599
Length=1584
Score = 36.6 bits (83), Expect = 0.071, Method: Composition-based stats.
Identities = 23/77 (29%), Positives = 37/77 (48%), Gaps = 7/77 (9%)
Query 161 ECEEGVSSPSPPP----SSRHLACQTFSAVYEAEEEPDAQVVTSVGSVLKSSDQPTDENA 216
+C+ G S+P+PPP R + + S ++ E D++ V +V + D P +
Sbjct 1508 DCQSGRSTPAPPPDDDNQKREITEEISSVNVDSNNEEDSESVLDEDTV--TGDVPVQHDH 1565
Query 217 PAEAEPASASPSADAGE 233
P E P PS+DA E
Sbjct 1566 PLE-RPTKLDPSSDAVE 1581
> CE20600
Length=1586
Score = 36.6 bits (83), Expect = 0.072, Method: Composition-based stats.
Identities = 23/77 (29%), Positives = 37/77 (48%), Gaps = 7/77 (9%)
Query 161 ECEEGVSSPSPPP----SSRHLACQTFSAVYEAEEEPDAQVVTSVGSVLKSSDQPTDENA 216
+C+ G S+P+PPP R + + S ++ E D++ V +V + D P +
Sbjct 1510 DCQSGRSTPAPPPDDDNQKREITEEISSVNVDSNNEEDSESVLDEDTV--TGDVPVQHDH 1567
Query 217 PAEAEPASASPSADAGE 233
P E P PS+DA E
Sbjct 1568 PLE-RPTKLDPSSDAVE 1583
> CE23607
Length=1449
Score = 36.2 bits (82), Expect = 0.097, Method: Composition-based stats.
Identities = 23/77 (29%), Positives = 37/77 (48%), Gaps = 7/77 (9%)
Query 161 ECEEGVSSPSPPP----SSRHLACQTFSAVYEAEEEPDAQVVTSVGSVLKSSDQPTDENA 216
+C+ G S+P+PPP R + + S ++ E D++ V +V + D P +
Sbjct 1373 DCQSGRSTPAPPPDDDNQKREITEEISSVNVDSNNEEDSESVLDEDTV--TGDVPVQHDH 1430
Query 217 PAEAEPASASPSADAGE 233
P E P PS+DA E
Sbjct 1431 PLE-RPTKLDPSSDAVE 1446
> Hs10947054
Length=1872
Score = 31.6 bits (70), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 23/59 (38%), Positives = 26/59 (44%), Gaps = 0/59 (0%)
Query 124 LCTVQHQQKEEGQEEVEEEEQQPQPIVEEPSSDTDASECEEGVSSPSPPPSSRHLACQT 182
LCT QH+QKEE E E PIV E S ++GV SS L QT
Sbjct 1601 LCTAQHKQKEEQAVSKESETCDHPPIVSEEDISVGYSTFQDGVPKTEGDSSSTALFPQT 1659
> YKR058w
Length=480
Score = 30.0 bits (66), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 32/115 (27%), Positives = 47/115 (40%), Gaps = 11/115 (9%)
Query 88 RLGYYVVYLTSVRAMKPFVR--HLLPTHPMPHML----DFISLCTVQHQQKEEGQEEVEE 141
LGY ++ KP ++ H + H P L +FI H Q E EE +E
Sbjct 91 NLGYQSS--PAMNYFKPSIKLIHFIGKH-KPWSLWSQKNFIK--NEYHDQWNEVYEEFKE 145
Query 142 EEQQPQPIVEEPSSDTDASECEEGVSSPSPPPSSRHLACQTFSAVYEAEEEPDAQ 196
E Q + + SD+D +E E ++ PPS+ Q + EE D Q
Sbjct 146 EHQLNNEVSKPKISDSDKTETPETITPVDAPPSNEPTTNQEIDTISTVEENVDNQ 200
> Hs22054104
Length=181
Score = 29.6 bits (65), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 27/85 (31%), Positives = 44/85 (51%), Gaps = 7/85 (8%)
Query 108 HLLPTHPMPHMLDFISLCTVQHQQKEEGQEEVEEEEQQPQPIVEEPSSDTDASECEEGVS 167
HL+ P P +LD S + K+ GQ ++ Q +P ++ P T + + ++GVS
Sbjct 8 HLM-GGPTPAVLD--SSSATWGRNKDRGQRAPSHQKAQEEPAMQ-PDRSTVSRKSDKGVS 63
Query 168 SPSPPPSSRHLACQTFSAVYEAEEE 192
SP+P P QT +A+ E E+E
Sbjct 64 SPNPAPLQHQ---QTEAALQEVEQE 85
Lambda K H
0.313 0.128 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8305360616
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40