bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1098_orf2
Length=223
Score E
Sequences producing significant alignments: (Bits) Value
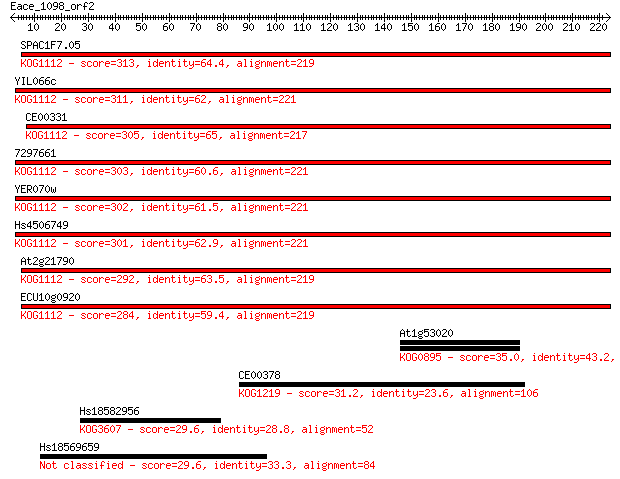
SPAC1F7.05 313 2e-85
YIL066c 311 1e-84
CE00331 305 6e-83
7297661 303 2e-82
YER070w 302 3e-82
Hs4506749 301 6e-82
At2g21790 292 4e-79
ECU10g0920 284 9e-77
At1g53020 35.0 0.13
CE00378 31.2 1.7
Hs18582956 29.6 4.9
Hs18569659 29.6 5.1
> SPAC1F7.05
Length=811
Score = 313 bits (802), Expect = 2e-85, Method: Compositional matrix adjust.
Identities = 141/219 (64%), Positives = 181/219 (82%), Gaps = 3/219 (1%)
Query 5 QMLWFAILQAQIETGTPYMLYKDACNRKSNHQHLGTLRSSNLCCEVVQYTSPEEVAVCNL 64
Q +W+AILQ+Q+ETG P+MLYKD+CNRKSN +++GT+R SNLC E+V+Y+SP+EVAVCNL
Sbjct 387 QKVWYAILQSQVETGNPFMLYKDSCNRKSNQKNVGTIRCSNLCTEIVEYSSPDEVAVCNL 446
Query 65 ASVSLPRFVDAEKKTFDYDKLKDVIKVMTRNLNNIIDRNYYPIPEARRSNLRHRPIGLGV 124
ASV+LP F+ K +++ KL DV+KV+TRNLN IID NYYP+PEARRSN+RHRP+GLGV
Sbjct 447 ASVALPTFIKDGK--YNFQKLHDVVKVVTRNLNKIIDVNYYPVPEARRSNMRHRPVGLGV 504
Query 125 QGLADAFLLLRLPFDSPAARDINKNIFECIYFAACEASMELAAIHGTYETYEGSPMSKGV 184
QGLADAF LRLPF+S A+ +N IFE IY AA EAS E+A + GTYE+YEGSP S+G+
Sbjct 505 QGLADAFFALRLPFESAGAKKLNIQIFETIYHAALEASCEIAQVEGTYESYEGSPASQGI 564
Query 185 FQFDMWGVKPDSGLCDWQGLRQKVKKYGLRNSLLVSPMP 223
Q+DMW V P + L DW L++K+ K+G+RNSLLV+PMP
Sbjct 565 LQYDMWNVNP-TDLWDWAELKEKIAKHGIRNSLLVAPMP 602
> YIL066c
Length=869
Score = 311 bits (796), Expect = 1e-84, Method: Compositional matrix adjust.
Identities = 137/225 (60%), Positives = 180/225 (80%), Gaps = 5/225 (2%)
Query 3 RRQMLWFAILQAQIETGTPYMLYKDACNRKSNHQHLGTLRSSNLCCEVVQYTSPEEVAVC 62
+ Q LW+AILQAQ ETGTP+M+YKDACNRK+N Q+LGT++SSNLCCE+V+Y+SP+E AVC
Sbjct 384 KAQKLWYAILQAQTETGTPFMVYKDACNRKTNQQNLGTIKSSNLCCEIVEYSSPDETAVC 443
Query 63 NLASVSLPRFV----DAEKKTFDYDKLKDVIKVMTRNLNNIIDRNYYPIPEARRSNLRHR 118
NLAS++LP FV D + ++++++L ++ KV+T NLN +IDRNYYP+PEAR SN++HR
Sbjct 444 NLASIALPAFVEVSEDGKTASYNFERLHEIAKVITHNLNRVIDRNYYPVPEARNSNMKHR 503
Query 119 PIGLGVQGLADAFLLLRLPFDSPAARDINKNIFECIYFAACEASMELAAIHGTYETYEGS 178
PI LGVQGLAD +++LRLPF+S A+ +NK IFE IY A EAS ELA G Y T+EGS
Sbjct 504 PIALGVQGLADTYMMLRLPFESEEAQTLNKQIFETIYHATLEASCELAQKEGKYSTFEGS 563
Query 179 PMSKGVFQFDMWGVKPDSGLCDWQGLRQKVKKYGLRNSLLVSPMP 223
P SKG+ QFDMW KP G+ DW+ LR+ + K+GLRNSL ++PMP
Sbjct 564 PASKGILQFDMWNAKP-FGMWDWETLRKDIVKHGLRNSLTMAPMP 607
> CE00331
Length=788
Score = 305 bits (780), Expect = 6e-83, Method: Compositional matrix adjust.
Identities = 141/217 (64%), Positives = 175/217 (80%), Gaps = 2/217 (0%)
Query 7 LWFAILQAQIETGTPYMLYKDACNRKSNHQHLGTLRSSNLCCEVVQYTSPEEVAVCNLAS 66
LW I+ QIETG PY+ YKDA NRKSN Q+LGT++ SNLC E+++Y++P+E+AVCNLAS
Sbjct 395 LWEHIVSNQIETGLPYITYKDAANRKSNQQNLGTIKCSNLCTEIIEYSAPDEIAVCNLAS 454
Query 67 VSLPRFVDAEKKTFDYDKLKDVIKVMTRNLNNIIDRNYYPIPEARRSNLRHRPIGLGVQG 126
++L R+V EKK FD+ KL +V KV+TRNLN IID NYYP+ EAR SN+RHRPIGLGVQG
Sbjct 455 IALNRYVTPEKK-FDFVKLAEVTKVITRNLNKIIDVNYYPVEEARNSNMRHRPIGLGVQG 513
Query 127 LADAFLLLRLPFDSPAARDINKNIFECIYFAACEASMELAAIHGTYETYEGSPMSKGVFQ 186
LAD F+L+R PF S ARD+NK IFE IY+AA EAS ELA ++G Y TYEGSP+SKG Q
Sbjct 514 LADCFMLMRYPFTSAEARDLNKRIFETIYYAALEASCELAELNGPYSTYEGSPVSKGQLQ 573
Query 187 FDMWGVKPDSGLCDWQGLRQKVKKYGLRNSLLVSPMP 223
FDMWGV P + CDW LR+K+ K+G+RNSLL++PMP
Sbjct 574 FDMWGVTP-TDQCDWATLRKKIAKHGIRNSLLMAPMP 609
> 7297661
Length=771
Score = 303 bits (776), Expect = 2e-82, Method: Compositional matrix adjust.
Identities = 134/221 (60%), Positives = 180/221 (81%), Gaps = 2/221 (0%)
Query 3 RRQMLWFAILQAQIETGTPYMLYKDACNRKSNHQHLGTLRSSNLCCEVVQYTSPEEVAVC 62
+ Q LWFAI++AQ+ETG PYML+KDACNRKSN Q++GT++ SNLC E+V+Y++P+E+AVC
Sbjct 355 KAQSLWFAIIEAQVETGNPYMLFKDACNRKSNQQNVGTIKCSNLCTEIVEYSAPDEIAVC 414
Query 63 NLASVSLPRFVDAEKKTFDYDKLKDVIKVMTRNLNNIIDRNYYPIPEARRSNLRHRPIGL 122
NLAS++L FV EK T+D+ KLK+V K++T+NLN IID NYYP+PEAR+SNLRHRP+G+
Sbjct 415 NLASIALNMFVTPEK-TYDFKKLKEVTKIVTKNLNKIIDINYYPLPEARKSNLRHRPVGI 473
Query 123 GVQGLADAFLLLRLPFDSPAARDINKNIFECIYFAACEASMELAAIHGTYETYEGSPMSK 182
G+QG ADA +L+R P++S A +N+ IFE IY+ A EAS ELA G YETYEGSP+SK
Sbjct 474 GIQGFADALILMRFPYESEEAGLLNQQIFETIYYGALEASCELAQTEGPYETYEGSPVSK 533
Query 183 GVFQFDMWGVKPDSGLCDWQGLRQKVKKYGLRNSLLVSPMP 223
G+ Q+DMW P + L DWQ L++ ++ +G+RNSLLV+PMP
Sbjct 534 GILQYDMWDKVP-TNLWDWQKLKESIRMHGVRNSLLVAPMP 573
> YER070w
Length=888
Score = 302 bits (774), Expect = 3e-82, Method: Compositional matrix adjust.
Identities = 136/225 (60%), Positives = 179/225 (79%), Gaps = 5/225 (2%)
Query 3 RRQMLWFAILQAQIETGTPYMLYKDACNRKSNHQHLGTLRSSNLCCEVVQYTSPEEVAVC 62
+ Q LW++IL+AQ ETGTP+++YKDACNRKSN ++LG ++SSNLCCE+V+Y++P+E AVC
Sbjct 384 KAQKLWYSILEAQTETGTPFVVYKDACNRKSNQKNLGVIKSSNLCCEIVEYSAPDETAVC 443
Query 63 NLASVSLPRFV----DAEKKTFDYDKLKDVIKVMTRNLNNIIDRNYYPIPEARRSNLRHR 118
NLASV+LP F+ D + T+++ KL ++ KV+TRNLN +IDRNYYP+ EAR+SN+RHR
Sbjct 444 NLASVALPAFIETSEDGKTSTYNFKKLHEIAKVVTRNLNRVIDRNYYPVEEARKSNMRHR 503
Query 119 PIGLGVQGLADAFLLLRLPFDSPAARDINKNIFECIYFAACEASMELAAIHGTYETYEGS 178
PI LGVQGLAD F+LLRLPFDS AR +N IFE IY A+ EAS ELA G YET++GS
Sbjct 504 PIALGVQGLADTFMLLRLPFDSEEARLLNIQIFETIYHASMEASCELAQKDGPYETFQGS 563
Query 179 PMSKGVFQFDMWGVKPDSGLCDWQGLRQKVKKYGLRNSLLVSPMP 223
P S+G+ QFDMW KP G+ DW LR+ + K+G+RNSL ++PMP
Sbjct 564 PASQGILQFDMWDQKP-YGMWDWDTLRKDIMKHGVRNSLTMAPMP 607
> Hs4506749
Length=792
Score = 301 bits (771), Expect = 6e-82, Method: Compositional matrix adjust.
Identities = 139/221 (62%), Positives = 179/221 (80%), Gaps = 2/221 (0%)
Query 3 RRQMLWFAILQAQIETGTPYMLYKDACNRKSNHQHLGTLRSSNLCCEVVQYTSPEEVAVC 62
+ Q LW+AI+++Q ETGTPYMLYKD+CNRKSN Q+LGT++ SNLC E+V+YTS +EVAVC
Sbjct 385 KAQQLWYAIIESQTETGTPYMLYKDSCNRKSNQQNLGTIKCSNLCTEIVEYTSKDEVAVC 444
Query 63 NLASVSLPRFVDAEKKTFDYDKLKDVIKVMTRNLNNIIDRNYYPIPEARRSNLRHRPIGL 122
NLAS++L +V +E T+D+ KL +V KV+ RNLN IID NYYP+PEA SN RHRPIG+
Sbjct 445 NLASLALNMYVTSEH-TYDFKKLAEVTKVVVRNLNKIIDINYYPVPEACLSNKRHRPIGI 503
Query 123 GVQGLADAFLLLRLPFDSPAARDINKNIFECIYFAACEASMELAAIHGTYETYEGSPMSK 182
GVQGLADAF+L+R PF+S A+ +NK IFE IY+ A EAS +LA G YETYEGSP+SK
Sbjct 504 GVQGLADAFILMRYPFESAEAQLLNKQIFETIYYGALEASCDLAKEQGPYETYEGSPVSK 563
Query 183 GVFQFDMWGVKPDSGLCDWQGLRQKVKKYGLRNSLLVSPMP 223
G+ Q+DMW V P + L DW+ L++K+ KYG+RNSLL++PMP
Sbjct 564 GILQYDMWNVTP-TDLWDWKVLKEKIAKYGIRNSLLIAPMP 603
> At2g21790
Length=816
Score = 292 bits (747), Expect = 4e-79, Method: Compositional matrix adjust.
Identities = 139/237 (58%), Positives = 175/237 (73%), Gaps = 19/237 (8%)
Query 5 QMLWFAILQAQIETGTPYMLYKDACNRKSNHQHLGTLRSSNLCCEVVQYTSPEEVAVCNL 64
Q LW+ IL +Q+ETGTPYML+KD+CNRKSN Q+LGT++SSNLC E+++YTSP E AVCNL
Sbjct 387 QQLWYEILTSQVETGTPYMLFKDSCNRKSNQQNLGTIKSSNLCTEIIEYTSPTETAVCNL 446
Query 65 ASVSLPRFV------------------DAEKKTFDYDKLKDVIKVMTRNLNNIIDRNYYP 106
AS++LPRFV D++ + FD++KL +V +T NLN IID NYYP
Sbjct 447 ASIALPRFVREKGVPLDSHPPKLAGSLDSKNRYFDFEKLAEVTATVTVNLNKIIDVNYYP 506
Query 107 IPEARRSNLRHRPIGLGVQGLADAFLLLRLPFDSPAARDINKNIFECIYFAACEASMELA 166
+ A+ SN+RHRPIG+GVQGLADAF+LL +PFDSP A+ +NK+IFE IY+ A +AS ELA
Sbjct 507 VETAKTSNMRHRPIGIGVQGLADAFILLGMPFDSPEAQQLNKDIFETIYYHALKASTELA 566
Query 167 AIHGTYETYEGSPMSKGVFQFDMWGVKPDSGLCDWQGLRQKVKKYGLRNSLLVSPMP 223
A G YETY GSP+SKG+ Q DMW V P S DW LR + K G+RNSLLV+PMP
Sbjct 567 ARLGPYETYAGSPVSKGILQPDMWNVIP-SDRWDWAVLRDMISKNGVRNSLLVAPMP 622
> ECU10g0920
Length=768
Score = 284 bits (727), Expect = 9e-77, Method: Compositional matrix adjust.
Identities = 130/219 (59%), Positives = 175/219 (79%), Gaps = 3/219 (1%)
Query 5 QMLWFAILQAQIETGTPYMLYKDACNRKSNHQHLGTLRSSNLCCEVVQYTSPEEVAVCNL 64
Q LW AI++AQIETGTPYM YKDACNR SN QHLGT++SSNLC E+V+Y+S EE +VCNL
Sbjct 380 QKLWKAIIEAQIETGTPYMCYKDACNRLSNQQHLGTIKSSNLCAEIVEYSSGEETSVCNL 439
Query 65 ASVSLPRFVDAEKKTFDYDKLKDVIKVMTRNLNNIIDRNYYPIPEARRSNLRHRPIGLGV 124
AS+ LP FV + FD++ + V+K++T NLN +ID NYYP+ EARRSN+R+RPIG+GV
Sbjct 440 ASICLPMFV--KDGWFDFEAFRRVVKILTVNLNRVIDFNYYPVEEARRSNMRNRPIGIGV 497
Query 125 QGLADAFLLLRLPFDSPAARDINKNIFECIYFAACEASMELAAIHGTYETYEGSPMSKGV 184
QGLAD F +LRL F+S AR +N++IFE +Y++A EAS ELA G + +YEGSP+SKG+
Sbjct 498 QGLADLFAILRLAFESDGARSLNQDIFEAMYYSAMEASCELAEKEGPFPSYEGSPISKGI 557
Query 185 FQFDMWGVKPDSGLCDWQGLRQKVKKYGLRNSLLVSPMP 223
F F++ G K SG DW+GLR++++++G+RNSLL++ MP
Sbjct 558 FHFELAGRKA-SGNWDWEGLRERIRRHGVRNSLLIALMP 595
> At1g53020
Length=1163
Score = 35.0 bits (79), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 19/46 (41%), Positives = 28/46 (60%), Gaps = 6/46 (13%)
Query 146 INKNIFECIYFAACEASMEL--AAIHGTYETYEGSPMSKGVFQFDM 189
+N+N+ E I+ ACE+ M+L A I G EG+P G+F FD+
Sbjct 929 LNQNLPETIFVRACESRMDLLRAVIIGA----EGTPYHDGLFFFDI 970
Score = 32.7 bits (73), Expect = 0.55, Method: Compositional matrix adjust.
Identities = 18/46 (39%), Positives = 28/46 (60%), Gaps = 6/46 (13%)
Query 146 INKNIFECIYFAACEASMEL--AAIHGTYETYEGSPMSKGVFQFDM 189
++KN+ E I+ ACE+ ++L A I G EG+P G+F FD+
Sbjct 617 LDKNLPETIFVRACESRIDLLRAVIIGA----EGTPYHDGLFFFDI 658
> CE00378
Length=3343
Score = 31.2 bits (69), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 25/106 (23%), Positives = 53/106 (50%), Gaps = 14/106 (13%)
Query 86 KDVIKVMTRNLNNIIDRNYYPIPEARRSNLRHRPIGLGVQGLADAFLLLRLPFDSPAARD 145
+ +I ++ NL+ DRN +P+ AR+ +R + + L ++L +PFDSP
Sbjct 2460 RKIINLVVNNLD--ADRNSFPL--ARQPTIR-KSMSLP------KTMILNIPFDSPTGTI 2508
Query 146 INKNIFECIYFAACEASMELAAIHGTYETYEGSPMSKGVFQFDMWG 191
I KN+ + + + ++ + +G+ +P+ + Q D++G
Sbjct 2509 IWKNLENAVQYMENQKNVNFS--NGSKNLILKTPLEE-TMQIDIFG 2551
> Hs18582956
Length=493
Score = 29.6 bits (65), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 15/52 (28%), Positives = 25/52 (48%), Gaps = 0/52 (0%)
Query 27 DACNRKSNHQHLGTLRSSNLCCEVVQYTSPEEVAVCNLASVSLPRFVDAEKK 78
D C S LG++ ++ LCC +Y +P V LP++ D +K+
Sbjct 386 DRCCETSCTLSLGSVCNTGLCCHKCKYAAPGVVCRDLGGICDLPKYCDGKKE 437
> Hs18569659
Length=859
Score = 29.6 bits (65), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 28/93 (30%), Positives = 38/93 (40%), Gaps = 10/93 (10%)
Query 12 LQAQIETGTPYMLYKDACNRKSNHQHLGTLRSSNLCCEVVQYTS----PEEVAVCNL--- 64
++ + E GT MLY D N QH GT++ + C T P E + NL
Sbjct 316 IRCEKEAGTSLMLYLDENRNNRNSQH-GTIKETRSDCLDFSSTELLLLPYETTIRNLQVQ 374
Query 65 ASVSLPRFVDAEKKTFDYDKLKDVIK--VMTRN 95
+ SL + Y+KL D K TRN
Sbjct 375 VTASLGHSIQGTLTDVFYEKLADHFKNPQQTRN 407
Lambda K H
0.322 0.137 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4255059914
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40