bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1122_orf1
Length=166
Score E
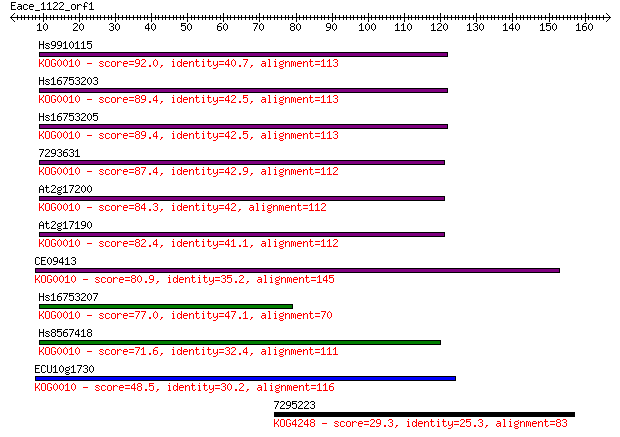
Sequences producing significant alignments: (Bits) Value
Hs9910115 92.0 5e-19
Hs16753203 89.4 3e-18
Hs16753205 89.4 3e-18
7293631 87.4 1e-17
At2g17200 84.3 1e-16
At2g17190 82.4 3e-16
CE09413 80.9 1e-15
Hs16753207 77.0 1e-14
Hs8567418 71.6 7e-13
ECU10g1730 48.5 6e-06
7295223 29.3 3.5
> Hs9910115
Length=601
Score = 92.0 bits (227), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 46/113 (40%), Positives = 72/113 (63%), Gaps = 5/113 (4%)
Query 9 LREQNPEINSLLSDPQLFRMGMDAMRNPSLLREMIRNTDRALASIEAVPGGFNALTRVCR 68
L E+NPEI+ +L++P+L R M+ RNP++++EM+RN DRAL+++E++PGG+NAL R+
Sbjct 228 LMERNPEISHMLNNPELMRQTMELARNPAMMQEMMRNQDRALSNLESIPGGYNALRRMYT 287
Query 69 GVQEPMHEAAIQIDAELRGGTARGERGNPTGEATNRRRDSGPSKEALPNPWAP 121
+QEPM AA + GN ++ R ++E LPNPW+P
Sbjct 288 DIQEPMFSAA---QEQFGNNPFSSLAGNSDSSSSQPLRTE--NREPLPNPWSP 335
> Hs16753203
Length=589
Score = 89.4 bits (220), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 48/118 (40%), Positives = 74/118 (62%), Gaps = 14/118 (11%)
Query 9 LREQNPEINSLLSDPQLFRMGMDAMRNPSLLREMIRNTDRALASIEAVPGGFNALTRVCR 68
L ++NPEI+ +L++P + R ++ RNP++++EM+RN DRAL+++E++PGG+NAL R+
Sbjct 218 LIQRNPEISHMLNNPDIMRQTLELARNPAMMQEMMRNQDRALSNLESIPGGYNALRRMYT 277
Query 69 GVQEPMHEAAIQ-----IDAELRGGTARGERGNPTGEATNRRRDSGPSKEALPNPWAP 121
+QEPM AA + A L T+ GE P+ R + P LPNPWAP
Sbjct 278 DIQEPMLSAAQEQFGGNPFASLVSNTSSGEGSQPS-----RTENRDP----LPNPWAP 326
> Hs16753205
Length=561
Score = 89.4 bits (220), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 48/118 (40%), Positives = 74/118 (62%), Gaps = 14/118 (11%)
Query 9 LREQNPEINSLLSDPQLFRMGMDAMRNPSLLREMIRNTDRALASIEAVPGGFNALTRVCR 68
L ++NPEI+ +L++P + R ++ RNP++++EM+RN DRAL+++E++PGG+NAL R+
Sbjct 218 LIQRNPEISHMLNNPDIMRQTLELARNPAMMQEMMRNQDRALSNLESIPGGYNALRRMYT 277
Query 69 GVQEPMHEAAIQ-----IDAELRGGTARGERGNPTGEATNRRRDSGPSKEALPNPWAP 121
+QEPM AA + A L T+ GE P+ R + P LPNPWAP
Sbjct 278 DIQEPMLSAAQEQFGGNPFASLVSNTSSGEGSQPS-----RTENRDP----LPNPWAP 326
> 7293631
Length=547
Score = 87.4 bits (215), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 48/117 (41%), Positives = 70/117 (59%), Gaps = 13/117 (11%)
Query 9 LREQNPEINSLLSDPQLFRMGMDAMRNPSLLREMIRNTDRALASIEAVPGGFNALTRVCR 68
L ++NPEI+ +L++P L R M+ RNPS+L+E++R+ DRA++++E+VPGG++AL R+ R
Sbjct 171 LMQRNPEISHMLNNPDLLRQTMELARNPSMLQELMRSHDRAMSNLESVPGGYSALQRIYR 230
Query 69 GVQEPMHEAAIQI-----DAELRGGTARGERGNPTGEATNRRRDSGPSKEALPNPWA 120
+QEPM AA + A L G G NP NR LPNPW
Sbjct 231 DIQEPMMNAATESFGRNPFAGLVDGGGSGAGNNPQQGTENRN--------PLPNPWG 279
> At2g17200
Length=551
Score = 84.3 bits (207), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 47/126 (37%), Positives = 69/126 (54%), Gaps = 14/126 (11%)
Query 9 LREQNPEINSLLSDPQLFRMGMDAMRNPSLLREMIRNTDRALASIEAVPGGFNALTRVCR 68
L ++NPE+ +L+DP + R ++A RNP L+REM+RNTDRA+++IE++P GFN L R+
Sbjct 203 LVDRNPELGHVLNDPSILRQTLEAARNPELMREMMRNTDRAMSNIESMPEGFNMLRRMYE 262
Query 69 GVQEPMHEAAIQ------------IDAEL--RGGTARGERGNPTGEATNRRRDSGPSKEA 114
VQEP+ A A L +G T +G + N + P+
Sbjct 263 NVQEPLMNATTMSGNAGNNTGSNPFAALLGNQGVTTQGSDASNNSSTPNAGTGTIPNANP 322
Query 115 LPNPWA 120
LPNPW
Sbjct 323 LPNPWG 328
> At2g17190
Length=536
Score = 82.4 bits (202), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 46/125 (36%), Positives = 68/125 (54%), Gaps = 13/125 (10%)
Query 9 LREQNPEINSLLSDPQLFRMGMDAMRNPSLLREMIRNTDRALASIEAVPGGFNALTRVCR 68
L ++NPE+ +L+DP + R ++A RNP L+REM+RNTDRA+++IE++P GFN L R+
Sbjct 198 LVDRNPELGHVLNDPSILRQTLEAARNPELMREMMRNTDRAMSNIESMPEGFNMLRRMYE 257
Query 69 GVQEPMHEAAIQID-----------AELRGGTARGERGNPTGEATN--RRRDSGPSKEAL 115
VQEP+ A + A L G +G+ T + P+ L
Sbjct 258 NVQEPLMNATTMSENAGNNTSSNPFAALLGNQGVTTQGSDTSNNISAPNAETGTPNANPL 317
Query 116 PNPWA 120
PNPW
Sbjct 318 PNPWG 322
> CE09413
Length=502
Score = 80.9 bits (198), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 51/145 (35%), Positives = 81/145 (55%), Gaps = 12/145 (8%)
Query 8 GLREQNPEINSLLSDPQLFRMGMDAMRNPSLLREMIRNTDRALASIEAVPGGFNALTRVC 67
L E+NPE+ +L+DP + R M+ +RNP++ +EM+RN D+A+ +++ +PGG AL R+
Sbjct 166 ALIERNPEVGHILNDPNVMRQTMEMIRNPNMFQEMMRNHDQAIRNLQGIPGGEAALERLY 225
Query 68 RGVQEPMHEAAIQIDAELRGGTARGERGNPTGEATNRRRDSGPSKEALPNPWAPQQSQNS 127
VQEP+ +A L G RG+ + E R + EALPNPWA N+
Sbjct 226 NDVQEPLLNSATN---SLSGNPFASLRGDQSSEPR-VDRAGQENNEALPNPWA----SNA 277
Query 128 LQQQQQQQSSESAGEAHELNTYLEA 152
Q Q ++ SA + N+ L++
Sbjct 278 NQATNNQSNNRSA----DFNSLLDS 298
> Hs16753207
Length=624
Score = 77.0 bits (188), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 33/70 (47%), Positives = 54/70 (77%), Gaps = 0/70 (0%)
Query 9 LREQNPEINSLLSDPQLFRMGMDAMRNPSLLREMIRNTDRALASIEAVPGGFNALTRVCR 68
L ++NPEI+ LL++P + R ++ RNP++++EM+RN D AL+++E++PGG+NAL R+
Sbjct 214 LIQRNPEISHLLNNPDIMRQTLEIARNPAMMQEMMRNQDLALSNLESIPGGYNALRRMYT 273
Query 69 GVQEPMHEAA 78
+QEPM AA
Sbjct 274 DIQEPMLNAA 283
> Hs8567418
Length=655
Score = 71.6 bits (174), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 36/111 (32%), Positives = 62/111 (55%), Gaps = 4/111 (3%)
Query 9 LREQNPEINSLLSDPQLFRMGMDAMRNPSLLREMIRNTDRALASIEAVPGGFNALTRVCR 68
L + NPEI +L++P++ R ++ +RNP++++EMIR+ DR L+++E++PGG+N L +
Sbjct 200 LIQHNPEIGHILNNPEIMRQTLEFLRNPAMMQEMIRSQDRVLSNLESIPGGYNVLCTMYT 259
Query 69 GVQEPMHEAAIQIDAELRGGTARGERGNPTGEATNRRRDSGPSKEALPNPW 119
+ +PM A E GG T + + + LPNPW
Sbjct 260 DIMDPMLNAV----QEQFGGNPFATATTDNATTTTSQPSRMENCDPLPNPW 306
> ECU10g1730
Length=275
Score = 48.5 bits (114), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 35/120 (29%), Positives = 61/120 (50%), Gaps = 20/120 (16%)
Query 8 GLREQ---NPEINSLLSDPQL-FRMGMDAMRNPSLLREMIRNTDRALASIEAVPGGFNAL 63
GL+E+ NPE+ ++S L + M +M NP + ++N D ++ +E +PGG N +
Sbjct 115 GLKEEMNNNPELRMMMSSSNLQDELEMFSM-NPEYMNTQLKNLDITMSKLENIPGGLNMI 173
Query 64 TRVCRGVQEPMHEAAIQIDAELRGGTARGERGNPTGEATNRRRDSGPSKEALPNPWAPQQ 123
+ + + VQ+P+ A L+ G RG R R+ P +EA+P AP++
Sbjct 174 SSMIKDVQDPLSSA-------LKEGMGRGYR------VKEGRKIDKPIEEAIPG--APKE 218
> 7295223
Length=1199
Score = 29.3 bits (64), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 21/83 (25%), Positives = 36/83 (43%), Gaps = 1/83 (1%)
Query 74 MHEAAIQIDAELRGGTARGERGNPTGEATNRRRDSGPSKEALPNPWAPQQSQNSLQQQQQ 133
+ E + D E+ TA G +P E S P + PN W P +++ LQ Q +
Sbjct 1025 LQEQTLDTDVEMSEVTASGSNSSPADELPAVIVGSEPWHMSFPNDWLPVITRD-LQTQAE 1083
Query 134 QQSSESAGEAHELNTYLEARRRQ 156
Q + + + + A+RR+
Sbjct 1084 QSNRPQPPFSDAYISGMSAKRRK 1106
Lambda K H
0.311 0.125 0.350
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2389760076
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40