bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1177_orf1
Length=161
Score E
Sequences producing significant alignments: (Bits) Value
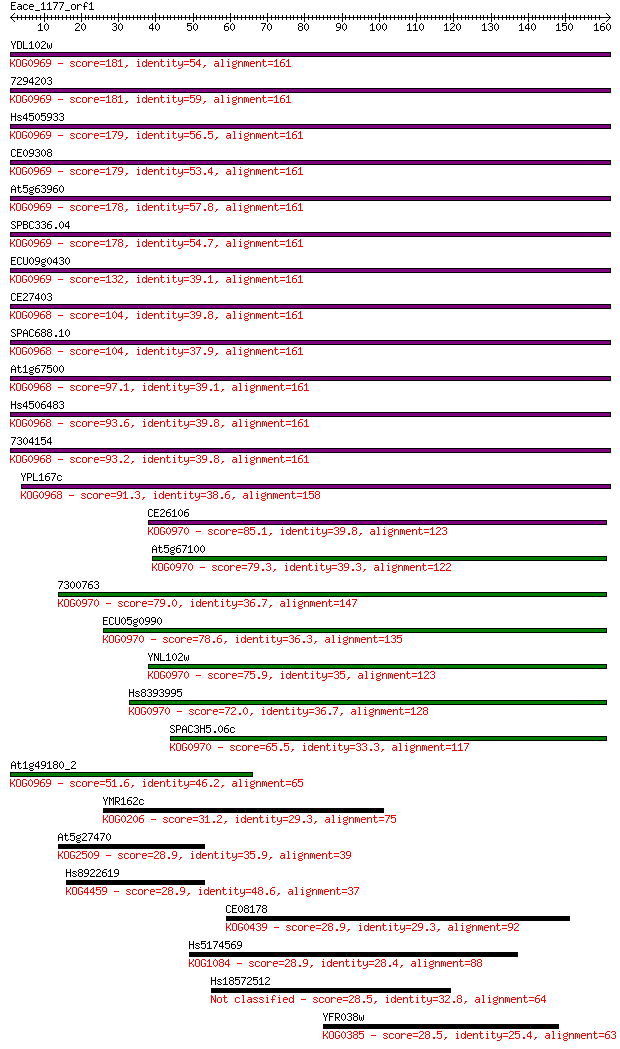
YDL102w 181 6e-46
7294203 181 6e-46
Hs4505933 179 2e-45
CE09308 179 2e-45
At5g63960 178 3e-45
SPBC336.04 178 4e-45
ECU09g0430 132 2e-31
CE27403 104 6e-23
SPAC688.10 104 7e-23
At1g67500 97.1 1e-20
Hs4506483 93.6 2e-19
7304154 93.2 2e-19
YPL167c 91.3 8e-19
CE26106 85.1 5e-17
At5g67100 79.3 3e-15
7300763 79.0 3e-15
ECU05g0990 78.6 5e-15
YNL102w 75.9 4e-14
Hs8393995 72.0 5e-13
SPAC3H5.06c 65.5 4e-11
At1g49180_2 51.6 7e-07
YMR162c 31.2 1.0
At5g27470 28.9 4.0
Hs8922619 28.9 4.0
CE08178 28.9 4.1
Hs5174569 28.9 4.4
Hs18572512 28.5 6.3
YFR038w 28.5 6.7
> YDL102w
Length=1097
Score = 181 bits (459), Expect = 6e-46, Method: Compositional matrix adjust.
Identities = 87/162 (53%), Positives = 117/162 (72%), Gaps = 3/162 (1%)
Query 1 LLTRGQQIKVTSQLLRKCRQLHYLVPSVKRVGGDRDDKYEGATVLEPLKGFYNMPISTLD 60
LL RGQQIKV SQL RKC ++ ++P+++ D D+YEGATV+EP++G+Y++PI+TLD
Sbjct 551 LLARGQQIKVVSQLFRKCLEIDTVIPNMQSQASD--DQYEGATVIEPIRGYYDVPIATLD 608
Query 61 FASLYPSIMMAHNICYSTLVKGTDVQTLQQDTLTHTTSSPR-HTFVKASVRRGVLPMIVE 119
F SLYPSIMMAHN+CY+TL V+ L +P FV RRG+LP+I++
Sbjct 609 FNSLYPSIMMAHNLCYTTLCNKATVERLNLKIDEDYVITPNGDYFVTTKRRRGILPIILD 668
Query 120 ELIAARKVAKAEMSKAEDKFMKSVLNGRQLALKITANSVYGY 161
ELI+ARK AK ++ +D F + VLNGRQLALKI+ANSVYG+
Sbjct 669 ELISARKRAKKDLRDEKDPFKRDVLNGRQLALKISANSVYGF 710
> 7294203
Length=1092
Score = 181 bits (459), Expect = 6e-46, Method: Compositional matrix adjust.
Identities = 95/167 (56%), Positives = 121/167 (72%), Gaps = 10/167 (5%)
Query 1 LLTRGQQIKVTSQLLRKCRQLHYLVPSVKRVGGDRDDKYEGATVLEPLKGFYNMPISTLD 60
LLTRGQQIKV SQLLRK + +++PS G D ++YEGATV+EP +G+Y PISTLD
Sbjct 530 LLTRGQQIKVLSQLLRKAKTKGFIMPSYTSQGSD--EQYEGATVIEPKRGYYADPISTLD 587
Query 61 FASLYPSIMMAHNICYSTLVKG------TDVQTLQQDTLTHTTSSPRHTFVKASVRRGVL 114
FASLYPSIMMAHN+CY+TLV G + LQ D + T ++ + FVK+ VRRG+L
Sbjct 588 FASLYPSIMMAHNLCYTTLVLGGTREKLRQQENLQDDQVERTPAN--NYFVKSEVRRGLL 645
Query 115 PMIVEELIAARKVAKAEMSKAEDKFMKSVLNGRQLALKITANSVYGY 161
P I+E L+AARK AK ++ D F + VL+GRQLALKI+ANSVYG+
Sbjct 646 PEILESLLAARKRAKNDLKVETDPFKRKVLDGRQLALKISANSVYGF 692
> Hs4505933
Length=1107
Score = 179 bits (455), Expect = 2e-45, Method: Compositional matrix adjust.
Identities = 91/163 (55%), Positives = 120/163 (73%), Gaps = 7/163 (4%)
Query 1 LLTRGQQIKVTSQLLRKCRQLHYLVPSVKRVGGDRDDKYEGATVLEPLKGFYNMPISTLD 60
LL+RGQQ+KV SQLLR+ L+P VK GG+ Y GATV+EPLKG+Y++PI+TLD
Sbjct 546 LLSRGQQVKVVSQLLRQAMHEGLLMPVVKSEGGE---DYTGATVIEPLKGYYDVPIATLD 602
Query 61 FASLYPSIMMAHNICYSTLVKGTDVQT--LQQDTLTHTTSSPRHTFVKASVRRGVLPMIV 118
F+SLYPSIMMAHN+CY+TL++ Q L +D T + FVK SVR+G+LP I+
Sbjct 603 FSSLYPSIMMAHNLCYTTLLRPGTAQKLGLTEDQFIRTPTG--DEFVKTSVRKGLLPQIL 660
Query 119 EELIAARKVAKAEMSKAEDKFMKSVLNGRQLALKITANSVYGY 161
E L++ARK AKAE++K D + VL+GRQLALK++ANSVYG+
Sbjct 661 ENLLSARKRAKAELAKETDPLRRQVLDGRQLALKVSANSVYGF 703
> CE09308
Length=1081
Score = 179 bits (453), Expect = 2e-45, Method: Compositional matrix adjust.
Identities = 86/161 (53%), Positives = 120/161 (74%), Gaps = 5/161 (3%)
Query 1 LLTRGQQIKVTSQLLRKCRQLHYLVPSVKRVGGDRDDKYEGATVLEPLKGFYNMPISTLD 60
LLT+GQQIK+ S +LR+C+Q ++ +P ++ GD + YEGATV++P++GFYN PI+TLD
Sbjct 516 LLTKGQQIKILSMMLRRCKQNNFFLPVIEANSGD-GEGYEGATVIDPIRGFYNEPIATLD 574
Query 61 FASLYPSIMMAHNICYSTLVKGTDVQTLQQDTLTHTTSSPRHTFVKASVRRGVLPMIVEE 120
FASLYPSIM+AHN+CY+TL+K Q ++ + T S F S RRG+LP I+E+
Sbjct 575 FASLYPSIMIAHNLCYTTLLKSP--QGVENEDYIRTPSG--QYFATKSKRRGLLPEILED 630
Query 121 LIAARKVAKAEMSKAEDKFMKSVLNGRQLALKITANSVYGY 161
++AARK AK +M +D+F + V NGRQLALKI+ANSVYG+
Sbjct 631 ILAARKRAKNDMKNEKDEFKRMVYNGRQLALKISANSVYGF 671
> At5g63960
Length=1081
Score = 178 bits (452), Expect = 3e-45, Method: Compositional matrix adjust.
Identities = 93/162 (57%), Positives = 121/162 (74%), Gaps = 3/162 (1%)
Query 1 LLTRGQQIKVTSQLLRKCRQLHYLVPSVKRVGGDRDDKYEGATVLEPLKGFYNMPISTLD 60
LL RGQ IKV SQLLRK +Q + ++P+ K+ G ++ YEGATVLE GFY PI+TLD
Sbjct 540 LLARGQSIKVLSQLLRKGKQKNLVLPNAKQSGSEQG-TYEGATVLEARTGFYEKPIATLD 598
Query 61 FASLYPSIMMAHNICYSTLVKGTDVQTLQQDTLTHTTSSPR-HTFVKASVRRGVLPMIVE 119
FASLYPSIMMA+N+CY TLV DV+ L H T +P TFVK ++++G+LP I+E
Sbjct 599 FASLYPSIMMAYNLCYCTLVTPEDVRKLNLPP-EHVTKTPSGETFVKQTLQKGILPEILE 657
Query 120 ELIAARKVAKAEMSKAEDKFMKSVLNGRQLALKITANSVYGY 161
EL+ ARK AKA++ +A+D K+VL+GRQLALKI+ANSVYG+
Sbjct 658 ELLTARKRAKADLKEAKDPLEKAVLDGRQLALKISANSVYGF 699
> SPBC336.04
Length=1086
Score = 178 bits (451), Expect = 4e-45, Method: Compositional matrix adjust.
Identities = 88/162 (54%), Positives = 122/162 (75%), Gaps = 3/162 (1%)
Query 1 LLTRGQQIKVTSQLLRKCRQLHYLVPSVKRVGGDRDDKYEGATVLEPLKGFYNMPISTLD 60
LL RGQQIKV SQL RK Q +VP++ RV G D++YEGATV+EP+KG+Y+ PI+TLD
Sbjct 530 LLARGQQIKVISQLFRKALQHDLVVPNI-RVNG-TDEQYEGATVIEPIKGYYDTPIATLD 587
Query 61 FASLYPSIMMAHNICYSTLVKGTDVQTLQ-QDTLTHTTSSPRHTFVKASVRRGVLPMIVE 119
F+SLYPSIM AHN+CY+TL+ + L+ + + ++ + FVK VR+G+LP+I+
Sbjct 588 FSSLYPSIMQAHNLCYTTLLDSNTAELLKLKQDVDYSVTPNGDYFVKPHVRKGLLPIILA 647
Query 120 ELIAARKVAKAEMSKAEDKFMKSVLNGRQLALKITANSVYGY 161
+L+ ARK AKA++ K D F K+VL+GRQLALK++ANSVYG+
Sbjct 648 DLLNARKKAKADLKKETDPFKKAVLDGRQLALKVSANSVYGF 689
> ECU09g0430
Length=974
Score = 132 bits (333), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 63/162 (38%), Positives = 103/162 (63%), Gaps = 9/162 (5%)
Query 1 LLTRGQQIKVTSQLLRKCRQLHYLVPSVKRVGGDRDDKYEGATVLEPLKGFYNMPISTLD 60
TRG IK+ + + R + +++P + ++ +EG V+EP KGFYN P+S +D
Sbjct 428 FFTRGMAIKIFTLVYRAASKEDFMIPDIDPFESNK--TFEGGFVIEPRKGFYNKPVSVMD 485
Query 61 FASLYPSIMMAHNICYSTLVKGTDVQTLQQDTLTHTTSSPR-HTFVKASVRRGVLPMIVE 119
F+SLYPSIM++HN+CY+TL+ T +Q + T +P + F A ++G+LP I+
Sbjct 486 FSSLYPSIMISHNLCYTTLL------TKEQYRILGGTKTPTGNYFCSAERKKGLLPRILT 539
Query 120 ELIAARKVAKAEMSKAEDKFMKSVLNGRQLALKITANSVYGY 161
+L+ +RK K E+ + +D +++ LNGRQLA K+ ANS+YG+
Sbjct 540 DLLTSRKRIKEELEREKDSALRACLNGRQLAFKLCANSLYGF 581
> CE27403
Length=1154
Score = 104 bits (260), Expect = 6e-23, Method: Compositional matrix adjust.
Identities = 64/189 (33%), Positives = 103/189 (54%), Gaps = 28/189 (14%)
Query 1 LLTRGQQIKVTSQLLRKCRQLHYLVPSVKRVGGDRDDKYEG-ATVLEPLKGFYNMPISTL 59
+ TRG Q++V S LLR +++++ PS+ + + E +LEP Y P+ L
Sbjct 599 VWTRGSQLRVESMLLRLAHRMNFVAPSITHLQRNMMGSPEQLQLILEPQSKVYFDPVIVL 658
Query 60 DFASLYPSIMMAHNICYST-------LVKGTDVQTLQQDTL------------------- 93
DF SLYPS+++A+N CYST LV+ D +++ +
Sbjct 659 DFQSLYPSMVIAYNYCYSTILGKIGNLVQMNDESRNREEIVLGAIKYHPSKDDIVKLVAY 718
Query 94 THTTSSPRHT-FVKASVRRGVLPMIVEELIAARKVAKAEMSKAEDKFMKSVLNGRQLALK 152
+SP + FVK S R GV+P+++ E++AAR + K+ M + ++K +K +L+ RQLALK
Sbjct 719 KEVCASPLASMFVKKSKREGVMPLLLREILAARIMVKSAMKRTKNKKLKRILDARQLALK 778
Query 153 ITANSVYGY 161
+ AN YGY
Sbjct 779 LVANVSYGY 787
> SPAC688.10
Length=1480
Score = 104 bits (260), Expect = 7e-23, Method: Compositional matrix adjust.
Identities = 61/178 (34%), Positives = 95/178 (53%), Gaps = 17/178 (9%)
Query 1 LLTRGQQIKVTSQLLRKCRQLHYLVPSVKRVGGDRDDKYEG-ATVLEPLKGFYNMPISTL 59
+++RG Q KV S + R + +Y+ PS + E V+EP YN P+ L
Sbjct 895 VISRGSQFKVESIMFRIAKPENYIFPSPSAKQVAEQNALEALPLVMEPKSDLYNNPVVVL 954
Query 60 DFASLYPSIMMAHNICYSTLVKGTDVQT--LQQDTLTHTTS-------------SPR-HT 103
DF SLYPSI++A+N+CYST + + ++ + H+++ SP +
Sbjct 955 DFQSLYPSIIIAYNLCYSTCLGPVKIVNGKVKLGFMFHSSNPNIVNLIKNDVYISPNGYA 1014
Query 104 FVKASVRRGVLPMIVEELIAARKVAKAEMSKAEDKFMKSVLNGRQLALKITANSVYGY 161
+VK +VR+ +L ++EELI R + K M + ++ VLN RQLALK+ AN YGY
Sbjct 1015 YVKENVRKSLLAKMLEELIETRNMVKRGMKDCDSDYVNKVLNSRQLALKLIANVTYGY 1072
> At1g67500
Length=1894
Score = 97.1 bits (240), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 63/182 (34%), Positives = 94/182 (51%), Gaps = 25/182 (13%)
Query 1 LLTRGQQIKVTSQLLRKCRQLHYLVPSVKRVGGDRDDKYEGAT-----VLEPLKGFYNMP 55
+L+RG Q +V S LLR +YL S G++ + A V+EP FY+ P
Sbjct 1303 VLSRGSQYRVESMLLRLAHTQNYLAIS----PGNQQVASQPAMECVPLVMEPESAFYDDP 1358
Query 56 ISTLDFASLYPSIMMAHNICYSTLVK----------GTDVQTLQQDT---LTHTTSSPRH 102
+ LDF SLYPS+++A+N+C+ST + G +L D L +P
Sbjct 1359 VIVLDFQSLYPSMIIAYNLCFSTCLGKLAHLKMNTLGVSSYSLDLDVLQDLNQILQTPNS 1418
Query 103 T-FVKASVRRGVLPMIVEELIAARKVAKAEMSK--AEDKFMKSVLNGRQLALKITANSVY 159
+V VRRG+LP ++EE+++ R + K M K + + + N RQLALK+ AN Y
Sbjct 1419 VMYVPPEVRRGILPRLLEEILSTRIMVKKAMKKLTPSEAVLHRIFNARQLALKLIANVTY 1478
Query 160 GY 161
GY
Sbjct 1479 GY 1480
> Hs4506483
Length=3052
Score = 93.6 bits (231), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 64/182 (35%), Positives = 98/182 (53%), Gaps = 22/182 (12%)
Query 1 LLTRGQQIKVTSQLLRKCRQLHYL--VPSVKRVGGDRDDKYEGATVLEPLKGFYNMPIST 58
+LTRG Q +V S +LR + ++Y+ PSV++ R + ++EP FY+ +
Sbjct 2476 VLTRGSQYRVESMMLRIAKPMNYIPVTPSVQQRSQMRAPQC-VPLIMEPESRFYSNSVLV 2534
Query 59 LDFASLYPSIMMAHNICYSTLVK-------------GTDVQTLQQDTLTHT----TSSPR 101
LDF SLYPSI++A+N C+ST + G + D L T SP
Sbjct 2535 LDFQSLYPSIVIAYNYCFSTCLGHVENLGKYDEFKFGCTSLRVPPDLLYQVRHDITVSPN 2594
Query 102 H-TFVKASVRRGVLPMIVEELIAARKVAKAEM-SKAEDKFMKSVLNGRQLALKITANSVY 159
FVK SVR+GVLP ++EE++ R + K M + +D+ + +L+ RQL LK+ AN +
Sbjct 2595 GVAFVKPSVRKGVLPRMLEEILKTRFMVKQSMKAYKQDRALSRMLDARQLGLKLIANVTF 2654
Query 160 GY 161
GY
Sbjct 2655 GY 2656
> 7304154
Length=1869
Score = 93.2 bits (230), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 64/184 (34%), Positives = 96/184 (52%), Gaps = 24/184 (13%)
Query 1 LLTRGQQIKVTSQLLR--KCRQLHYLVPSVKRVGGDRDDKYEGATVLEPLKGFYNMPIST 58
+L+RG Q +V S +LR K + L L PSV+ R +Y A ++EP FY P+
Sbjct 1285 VLSRGSQFRVESMMLRIAKPKNLVPLSPSVQARAHMRAPEY-LALIMEPQSRFYADPLIV 1343
Query 59 LDFASLYPSIMMAHNICYSTLVKGTD-----------------VQTLQQDTLTH---TTS 98
LDF SLYPS+++A+N C+ST + + + + Q L H T S
Sbjct 1344 LDFQSLYPSMIIAYNYCFSTCLGRVEHLGGSSPFEFGASQLRVSRQMLQKLLEHDLVTVS 1403
Query 99 SPRHTFVKASVRRGVLPMIVEELIAARKVAKAEMSKAED-KFMKSVLNGRQLALKITANS 157
FVK VR G+LP ++ E++ R++ K M +D ++ +L+ RQL LK+ AN
Sbjct 1404 PCGVVFVKREVREGILPRMLTEILDTRQMVKQSMKLHKDSSALQRILHSRQLGLKLMANV 1463
Query 158 VYGY 161
YGY
Sbjct 1464 TYGY 1467
> YPL167c
Length=1504
Score = 91.3 bits (225), Expect = 8e-19, Method: Compositional matrix adjust.
Identities = 61/182 (33%), Positives = 93/182 (51%), Gaps = 28/182 (15%)
Query 4 RGQQIKVTSQLLRKCRQLHYLVPSVKRVGGDRDDKYEGAT-----VLEPLKGFYNMPIST 58
RG Q KV S L+R C+ +++ S G +D + + A V+EP FY P+
Sbjct 918 RGSQFKVESFLIRICKSESFILLS----PGKKDVRKQKALECVPLVMEPESAFYKSPLIV 973
Query 59 LDFASLYPSIMMAHNICYSTLVKGTDVQTLQQDTLTHTTSS-PRH--------------- 102
LDF SLYPSIM+ +N CYST++ L ++ L + S PR+
Sbjct 974 LDFQSLYPSIMIGYNYCYSTMIGRVREINLTENNLGVSKFSLPRNILALLKNDVTIAPNG 1033
Query 103 -TFVKASVRRGVLPMIVEELIAARKVAKAEMSKAEDK--FMKSVLNGRQLALKITANSVY 159
+ K SVR+ L ++ +++ R + K M++ D +K +LN +QLALK+ AN Y
Sbjct 1034 VVYAKTSVRKSTLSKMLTDILDVRVMIKKTMNEIGDDNTTLKRLLNNKQLALKLLANVTY 1093
Query 160 GY 161
GY
Sbjct 1094 GY 1095
> CE26106
Length=1443
Score = 85.1 bits (209), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 49/123 (39%), Positives = 68/123 (55%), Gaps = 13/123 (10%)
Query 38 KYEGATVLEPLKGFYNMPISTLDFASLYPSIMMAHNICYSTLVKGTDVQTLQQDTLTHTT 97
+Y G VLEP KG Y I LDF SLYPSI+ +NICY+TL D
Sbjct 816 QYSGGLVLEPKKGLYETLILLLDFNSLYPSIIQEYNICYTTLEYSKDSD--------EQL 867
Query 98 SSPRHTFVKASVRRGVLPMIVEELIAARKVAKAEMSKAEDKFMKSVLNGRQLALKITANS 157
S P+ T ++ GVLP + +L+ R+ K+ M ++ K ++ RQ+ALK+TANS
Sbjct 868 SVPQSTDIE-----GVLPREIRKLVECRRDVKSLMKSERNEAKKKQMDIRQMALKLTANS 922
Query 158 VYG 160
+YG
Sbjct 923 MYG 925
> At5g67100
Length=1492
Score = 79.3 bits (194), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 48/122 (39%), Positives = 68/122 (55%), Gaps = 17/122 (13%)
Query 39 YEGATVLEPLKGFYNMPISTLDFASLYPSIMMAHNICYSTLVKGTDVQTLQQDTLTHTTS 98
Y G VLEP +G Y+ + LDF SLYPSI+ +NIC++T+ + D
Sbjct 897 YAGGLVLEPKRGLYDKYVLLLDFNSLYPSIIQEYNICFTTIPRSED-------------G 943
Query 99 SPRHTFVKASVRRGVLPMIVEELIAARKVAKAEMSKAEDKFMKSVLNGRQLALKITANSV 158
PR + +S G+LP ++E L++ RK K +M K E L+ RQ ALK+TANS+
Sbjct 944 VPR---LPSSQTPGILPKLMEHLVSIRKSVKLKMKK-ETGLKYWELDIRQQALKLTANSM 999
Query 159 YG 160
YG
Sbjct 1000 YG 1001
> 7300763
Length=1472
Score = 79.0 bits (193), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 54/168 (32%), Positives = 83/168 (49%), Gaps = 34/168 (20%)
Query 14 LLRKCRQLHYLVPSVKRV-----GGDRDD---------------KYEGATVLEPLKGFYN 53
LL + +Y+VP K V GD D Y G VLEP++G Y
Sbjct 814 LLHAFHEKNYIVPDKKPVSKRSGAGDTDATLSGADATMQTKKKAAYAGGLVLEPMRGLYE 873
Query 54 MPISTLDFASLYPSIMMAHNICYSTLVKGTDVQTLQQDTLTHTTSSPRHTFVKASVRRGV 113
+ +DF SLYPSI+ +NIC++T+ + D L TL + + P G+
Sbjct 874 KYVLLMDFNSLYPSIIQEYNICFTTVQQPVDADELP--TLPDSKTEP-----------GI 920
Query 114 LPMIVEELIAARKVAKAEMSKAE-DKFMKSVLNGRQLALKITANSVYG 160
LP+ ++ L+ +RK K M+ + ++ + RQ+ALK+TANS+YG
Sbjct 921 LPLQLKRLVESRKEVKKLMAAPDLSPELQMQYHIRQMALKLTANSMYG 968
> ECU05g0990
Length=956
Score = 78.6 bits (192), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 49/137 (35%), Positives = 69/137 (50%), Gaps = 12/137 (8%)
Query 26 PSVKRVGGDRDDKYEGATVLEPLKGFYNMPISTLDFASLYPSIMMAHNICYST--LVKGT 83
P+V R R+ KY G VL P GFY + LDF SLYPSI+ N+C+ST + +
Sbjct 394 PTVSR----REVKYSGGLVLPPQTGFYEDLVLLLDFNSLYPSIIQEFNVCFSTIGMFDRS 449
Query 84 DVQTLQQDTLTHTTSSPRHTFVKASVRRGVLPMIVEELIAARKVAKAEMSKAEDKFMKSV 143
+ + L T +++ RG LP I+E + RK K + +
Sbjct 450 ISGNMSSEQLVRLTEDAKNS------ERGFLPRILEGFVRRRKAVKDLLKQGGTPEETKA 503
Query 144 LNGRQLALKITANSVYG 160
L+ RQ ALK+TANS+YG
Sbjct 504 LDIRQKALKLTANSIYG 520
> YNL102w
Length=1468
Score = 75.9 bits (185), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 43/123 (34%), Positives = 65/123 (52%), Gaps = 12/123 (9%)
Query 38 KYEGATVLEPLKGFYNMPISTLDFASLYPSIMMAHNICYSTLVKGTDVQTLQQDTLTHTT 97
KY+G V EP KG + + +DF SLYPSI+ NIC++T+ + ++ +
Sbjct 842 KYQGGLVFEPEKGLHKNYVLVMDFNSLYPSIIQEFNICFTTVDRN-------KEDIDELP 894
Query 98 SSPRHTFVKASVRRGVLPMIVEELIAARKVAKAEMSKAEDKFMKSVLNGRQLALKITANS 157
S P + V +GVLP ++ L+ R+ K M D + + RQ ALK+TANS
Sbjct 895 SVP-----PSEVDQGVLPRLLANLVDRRREVKKVMKTETDPHKRVQCDIRQQALKLTANS 949
Query 158 VYG 160
+YG
Sbjct 950 MYG 952
> Hs8393995
Length=1462
Score = 72.0 bits (175), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 47/131 (35%), Positives = 71/131 (54%), Gaps = 8/131 (6%)
Query 33 GDRDDKYEGATVLEPLKGFYNMPISTLDFASLYPSIMMAHNICYSTLVK-GTDVQTLQQD 91
G + Y G VL+P GFY+ I LDF SLYPSI+ NIC++T+ + ++ Q + +D
Sbjct 833 GRKKGAYAGGLVLDPKVGFYDKFILLLDFNSLYPSIIQEFNICFTTVQRVASEAQKVTED 892
Query 92 TLTHTTSSPRHTFVKASVRRGVLPMIVEELIAARKVAKAEMSKAEDKFMKSVL--NGRQL 149
S+ G+LP + +L+ RK K ++ K +D +L + RQ
Sbjct 893 GEQEQIPE----LPDPSLEMGILPREIRKLVERRKQVK-QLMKQQDLNPDLILQYDIRQK 947
Query 150 ALKITANSVYG 160
ALK+TANS+YG
Sbjct 948 ALKLTANSMYG 958
> SPAC3H5.06c
Length=1405
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 39/118 (33%), Positives = 60/118 (50%), Gaps = 10/118 (8%)
Query 44 VLEPLKGFYNMPISTLDFASLYPSIMMAHNICYSTLVKGTDVQTLQQDTLTHTTSSPRHT 103
V EP KG Y I +DF SLYPSI+ +NIC++T+ D D+ +P
Sbjct 830 VFEPQKGLYETCILVMDFNSLYPSIIQEYNICFTTV----DRSPSNSDSDDQIPDTP--- 882
Query 104 FVKASVRRGVLPMIVEELIAARKVAKAEM-SKAEDKFMKSVLNGRQLALKITANSVYG 160
AS +G+ P ++ L+ R+ K + + + + +Q ALK+TANS+YG
Sbjct 883 --SASANQGIFPRLIANLVERRRQIKGLLKDNSATPTQRLQWDIQQQALKLTANSMYG 938
> At1g49180_2
Length=167
Score = 51.6 bits (122), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 30/65 (46%), Positives = 40/65 (61%), Gaps = 5/65 (7%)
Query 1 LLTRGQQIKVTSQLLRKCRQLHYLVPSVKRVGGDRDDKYEGATVLEPLKGFYNMPISTLD 60
L++RG+ IKV SQLLRK +Q + ++ ++ E VLE GFY PI+TLD
Sbjct 107 LISRGESIKVLSQLLRKAKQKTWFFQMLRSWNLNK----ELMKVLEARTGFYE-PIATLD 161
Query 61 FASLY 65
FASLY
Sbjct 162 FASLY 166
> YMR162c
Length=1656
Score = 31.2 bits (69), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 22/88 (25%), Positives = 43/88 (48%), Gaps = 13/88 (14%)
Query 26 PSVKRV-GGDRDDKYEGATVLEPLKGFYNMPISTLD---FASLYPSIMMAHN-------- 73
PS++ + G D + A+V+ P + F S+ D F YP+ + +
Sbjct 676 PSMRSLFGKDNSHLSKQASVISPSETFSENIKSSFDLIQFIQRYPTALFSQKAKFFFLSL 735
Query 74 -ICYSTLVKGTDVQTLQQDTLTHTTSSP 100
+C+S L K T +++ +D++ + +SSP
Sbjct 736 ALCHSCLPKKTHNESIGEDSIEYQSSSP 763
> At5g27470
Length=451
Score = 28.9 bits (63), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 14 LLRKCRQLHYLVPSVKRVGGDRDDKYEGATVLEPLKGFY 52
++ KC QL + +V G+ DDKY AT +PL ++
Sbjct 209 VMAKCAQLAQFDEELYKVTGEGDDKYLIATAEQPLCAYH 247
> Hs8922619
Length=527
Score = 28.9 bits (63), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 18/46 (39%), Positives = 26/46 (56%), Gaps = 9/46 (19%)
Query 16 RKCRQLHYLVPSVKRVG-GDR--------DDKYEGATVLEPLKGFY 52
+CR+LH + + VG G R ++K+EGATVL+ LK Y
Sbjct 293 EQCRELHSVASGIMLVGDGYRGKTSPHTPNEKFEGATVLKALKSGY 338
> CE08178
Length=221
Score = 28.9 bits (63), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 27/92 (29%), Positives = 46/92 (50%), Gaps = 4/92 (4%)
Query 59 LDFASLYPSIMMAHNICYSTLVKGTDVQTLQQDTLTHTTSSPRHTFVKASVRRGVLPMIV 118
+++++L P+ +A +I ST K T+ Q LQ T S+ T V+AS + P
Sbjct 127 MEYSALPPTPTVASSIKQSTKHKRTERQ-LQS---TQKMSTENITSVQASTTQQECPTQE 182
Query 119 EELIAARKVAKAEMSKAEDKFMKSVLNGRQLA 150
++ A VAK + +K ++ F K + LA
Sbjct 183 DDPTAPTPVAKPKKAKRKEVFFKDQDDDDTLA 214
> Hs5174569
Length=3969
Score = 28.9 bits (63), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 25/88 (28%), Positives = 37/88 (42%), Gaps = 13/88 (14%)
Query 49 KGFYNMPISTLDFASLYPSIMMAHNICYSTLVKGTDVQTLQQDTLTHTTSSPRHTFVKAS 108
KG N+ + LD PS+ +C S T T+ H+TSS +A
Sbjct 964 KGRGNLEKTNLDLGPTAPSLEKEKTLCLS---------TPSSSTVKHSTSSIGSMLAQAD 1014
Query 109 VRRGVLPMIVEELIAARKVAKAEMSKAE 136
LPM + + + K AKA++ K E
Sbjct 1015 ----KLPMTDKRVASLLKKAKAQLCKIE 1038
> Hs18572512
Length=768
Score = 28.5 bits (62), Expect = 6.3, Method: Composition-based stats.
Identities = 21/65 (32%), Positives = 30/65 (46%), Gaps = 1/65 (1%)
Query 55 PISTLDFASLYPSIMMAHNICYSTLVKGTDVQTLQQDTLTHTTSSPRH-TFVKASVRRGV 113
PIS L+ A L S + +I ++GTD + Q D T P+ T + AS V
Sbjct 664 PISFLNPARLRESPLFQQDIHLQANLEGTDTRRFQWDMTRATEHIPQDLTAITASAANPV 723
Query 114 LPMIV 118
P +V
Sbjct 724 KPCMV 728
> YFR038w
Length=778
Score = 28.5 bits (62), Expect = 6.7, Method: Composition-based stats.
Identities = 16/65 (24%), Positives = 33/65 (50%), Gaps = 2/65 (3%)
Query 85 VQTLQQDTLTHTTSSPRHTFVKASVRRGVLPMIVEELIAARK--VAKAEMSKAEDKFMKS 142
+ L D L S HT +K + R + +++ ++ ++ + M+ A++KF K+
Sbjct 429 LNKLINDELQKNLISNLHTILKPFLLRRLKKVVLANILPPKREYIINCPMTSAQEKFYKA 488
Query 143 VLNGR 147
LNG+
Sbjct 489 GLNGK 493
Lambda K H
0.320 0.133 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2208717646
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40