bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1186_orf1
Length=183
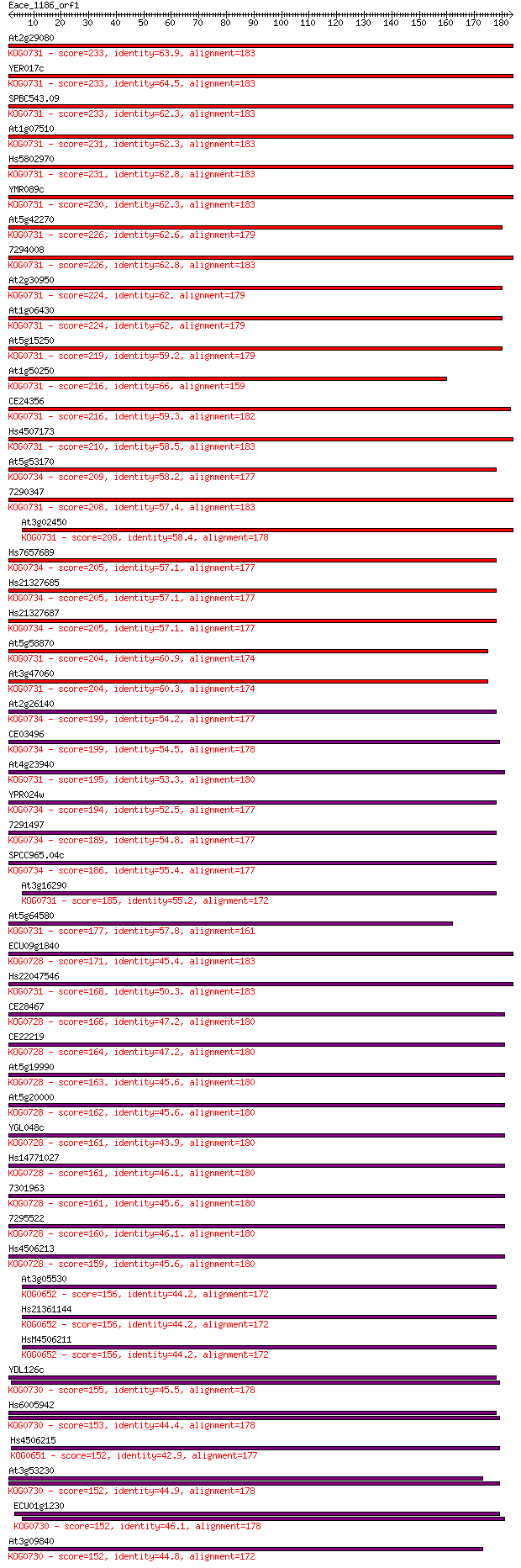
Score E
Sequences producing significant alignments: (Bits) Value
At2g29080 233 1e-61
YER017c 233 2e-61
SPBC543.09 233 2e-61
At1g07510 231 4e-61
Hs5802970 231 5e-61
YMR089c 230 1e-60
At5g42270 226 1e-59
7294008 226 2e-59
At2g30950 224 8e-59
At1g06430 224 1e-58
At5g15250 219 3e-57
At1g50250 216 2e-56
CE24356 216 2e-56
Hs4507173 210 1e-54
At5g53170 209 3e-54
7290347 208 4e-54
At3g02450 208 6e-54
Hs7657689 205 4e-53
Hs21327685 205 4e-53
Hs21327687 205 4e-53
At5g58870 204 9e-53
At3g47060 204 1e-52
At2g26140 199 3e-51
CE03496 199 3e-51
At4g23940 195 5e-50
YPR024w 194 6e-50
7291497 189 2e-48
SPCC965.04c 186 2e-47
At3g16290 185 4e-47
At5g64580 177 1e-44
ECU09g1840 171 1e-42
Hs22047546 168 6e-42
CE28467 166 2e-41
CE22219 164 9e-41
At5g19990 163 2e-40
At5g20000 162 2e-40
YGL048c 161 5e-40
Hs14771027 161 5e-40
7301963 161 7e-40
7295522 160 1e-39
Hs4506213 159 3e-39
At3g05530 156 2e-38
Hs21361144 156 2e-38
HsM4506211 156 2e-38
YDL126c 155 3e-38
Hs6005942 153 2e-37
Hs4506215 152 3e-37
At3g53230 152 4e-37
ECU01g1230 152 5e-37
At3g09840 152 5e-37
> At2g29080
Length=807
Score = 233 bits (595), Expect = 1e-61, Method: Compositional matrix adjust.
Identities = 117/185 (63%), Positives = 145/185 (78%), Gaps = 4/185 (2%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
KNP ++ +GAK+PKGALLVGPPGTGKTLLAKA AGE+GVPF SISGSDF+E+FVGVG S
Sbjct 340 KNPKKYEDLGAKIPKGALLVGPPGTGKTLLAKATAGESGVPFLSISGSDFMEMFVGVGPS 399
Query 61 RVRELFDEARKAAPSIIWIDEIDSV--GASRSTQFANSEREQTLNQLLVEMDGFSPHESV 118
RVR LF EAR+AAPSII+IDEID++ R N ERE TLNQLLVEMDGF V
Sbjct 400 RVRHLFQEARQAAPSIIFIDEIDAIGRARGRGGLGGNDERESTLNQLLVEMDGFGTTAGV 459
Query 119 VVLAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALAE 178
VVLAGTNR D+LD AL R GRFDR++ I++PD+K R +IFK++L+ ++L + + ++
Sbjct 460 VVLAGTNRPDILDKALLRPGRFDRQITIDKPDIKGRDQIFKIYLKKIKLDH--EPSYYSQ 517
Query 179 RMAAL 183
R+AAL
Sbjct 518 RLAAL 522
> YER017c
Length=761
Score = 233 bits (593), Expect = 2e-61, Method: Compositional matrix adjust.
Identities = 118/189 (62%), Positives = 144/189 (76%), Gaps = 6/189 (3%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
KNP + K+GAK+P+GA+L GPPGTGKTLLAKA AGEA VPF S+SGS+FVE+FVGVGAS
Sbjct 308 KNPGKYTKLGAKIPRGAILSGPPGTGKTLLAKATAGEANVPFLSVSGSEFVEMFVGVGAS 367
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASR----STQFANSEREQTLNQLLVEMDGFSPHE 116
RVR+LF +AR APSII+IDEID++G R + AN ERE TLNQLLVEMDGF+ +
Sbjct 368 RVRDLFTQARSMAPSIIFIDEIDAIGKERGKGGALGGANDEREATLNQLLVEMDGFTTSD 427
Query 117 SVVVLAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRV--DAN 174
VVVLAGTNR D+LD AL R GRFDR + I+ PDV R +I+ VHL+ L L P + D N
Sbjct 428 QVVVLAGTNRPDVLDNALMRPGRFDRHIQIDSPDVNGRQQIYLVHLKRLNLDPLLTDDMN 487
Query 175 ALAERMAAL 183
L+ ++A L
Sbjct 488 NLSGKLATL 496
> SPBC543.09
Length=773
Score = 233 bits (593), Expect = 2e-61, Method: Compositional matrix adjust.
Identities = 114/186 (61%), Positives = 147/186 (79%), Gaps = 5/186 (2%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
KNP ++++GAK+P+GA+L GPPGTGKTLLAKA AGEA VPF S+SGS+F+E+FVGVG S
Sbjct 316 KNPKFYERLGAKIPRGAILSGPPGTGKTLLAKATAGEANVPFLSVSGSEFLEMFVGVGPS 375
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASRST--QF-ANSEREQTLNQLLVEMDGFSPHES 117
RVR+LF ARK AP II+IDEID++G +R QF +N ERE TLNQLLVEMDGF+ E
Sbjct 376 RVRDLFATARKNAPCIIFIDEIDAIGKARGRGGQFGSNDERESTLNQLLVEMDGFTSSEH 435
Query 118 VVVLAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALA 177
+VV AGTNR D+LD AL R GRFDR++ I+RPD+ R +IFKVHL+ ++ + +D +A
Sbjct 436 IVVFAGTNRPDVLDPALLRPGRFDRQITIDRPDIGGREQIFKVHLKHIKAADNID--LIA 493
Query 178 ERMAAL 183
+R+A L
Sbjct 494 KRLAVL 499
> At1g07510
Length=748
Score = 231 bits (590), Expect = 4e-61, Method: Compositional matrix adjust.
Identities = 114/186 (61%), Positives = 145/186 (77%), Gaps = 5/186 (2%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
+NP ++ +GAK+PKGALLVGPPGTGKTLLAKA AGE+ VPF SISGSDF+E+FVGVG S
Sbjct 282 QNPKKYEDLGAKIPKGALLVGPPGTGKTLLAKATAGESAVPFLSISGSDFMEMFVGVGPS 341
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASRST---QFANSEREQTLNQLLVEMDGFSPHES 117
RVR LF EAR+ APSII+IDEID++G +R N ERE TLNQLLVEMDGF
Sbjct 342 RVRNLFQEARQCAPSIIFIDEIDAIGRARGRGGFSGGNDERESTLNQLLVEMDGFGTTAG 401
Query 118 VVVLAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALA 177
VVVLAGTNR D+LD AL R GRFDR++ I++PD+K R +IF+++L+ ++L + + +
Sbjct 402 VVVLAGTNRPDILDKALLRPGRFDRQITIDKPDIKGRDQIFQIYLKKIKLDH--EPSYYS 459
Query 178 ERMAAL 183
+R+AAL
Sbjct 460 QRLAAL 465
> Hs5802970
Length=797
Score = 231 bits (590), Expect = 5e-61, Method: Compositional matrix adjust.
Identities = 115/185 (62%), Positives = 145/185 (78%), Gaps = 2/185 (1%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
KNP +Q +GAK+PKGA+L GPPGTGKTLLAKA AGEA VPF ++SGS+F+E+FVGVG +
Sbjct 328 KNPKQYQDLGAKIPKGAILTGPPGTGKTLLAKATAGEANVPFITVSGSEFLEMFVGVGPA 387
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASRST-QF-ANSEREQTLNQLLVEMDGFSPHESV 118
RVR+LF ARK AP I++IDEID+VG R F SE+E TLNQLLVEMDGF+ +V
Sbjct 388 RVRDLFALARKNAPCILFIDEIDAVGRKRGRGNFGGQSEQENTLNQLLVEMDGFNTTTNV 447
Query 119 VVLAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALAE 178
V+LAGTNR D+LD AL R GRFDR++ I PD+K RA IFKVHL+PL+L ++ + LA
Sbjct 448 VILAGTNRPDILDPALLRPGRFDRQIFIGPPDIKGRASIFKVHLRPLKLDSTLEKDKLAR 507
Query 179 RMAAL 183
++A+L
Sbjct 508 KLASL 512
> YMR089c
Length=825
Score = 230 bits (587), Expect = 1e-60, Method: Compositional matrix adjust.
Identities = 114/186 (61%), Positives = 146/186 (78%), Gaps = 5/186 (2%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
K P+ ++KMGAK+P+GA+L GPPGTGKTLLAKA AGEAGVPF+ +SGS+FVE+FVGVGA+
Sbjct 368 KEPSRYEKMGAKIPRGAILSGPPGTGKTLLAKATAGEAGVPFYFVSGSEFVEMFVGVGAA 427
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASRST---QFANSEREQTLNQLLVEMDGFSPHES 117
RVR+LF AR+ APSI++IDEID++G +R AN ERE TLNQ+LVEMDGF+P +
Sbjct 428 RVRDLFKTARENAPSIVFIDEIDAIGKARQKGNFSGANDERENTLNQMLVEMDGFTPADH 487
Query 118 VVVLAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALA 177
VVVLAGTNR D+LD AL R GRFDR + I++P+++ R IF VHL L+L+ + L
Sbjct 488 VVVLAGTNRPDILDKALLRPGRFDRHINIDKPELEGRKAIFAVHLHHLKLAGEI--FDLK 545
Query 178 ERMAAL 183
R+AAL
Sbjct 546 NRLAAL 551
> At5g42270
Length=704
Score = 226 bits (577), Expect = 1e-59, Method: Compositional matrix adjust.
Identities = 112/181 (61%), Positives = 137/181 (75%), Gaps = 2/181 (1%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
KNP + +GAK+PKG LLVGPPGTGKTLLA+AVAGEAGVPFFS + S+FVELFVGVGAS
Sbjct 270 KNPDKYTALGAKIPKGCLLVGPPGTGKTLLARAVAGEAGVPFFSCAASEFVELFVGVGAS 329
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASRSTQF--ANSEREQTLNQLLVEMDGFSPHESV 118
RVR+LF++A+ AP I++IDEID+VG R N EREQT+NQLL EMDGFS + V
Sbjct 330 RVRDLFEKAKSKAPCIVFIDEIDAVGRQRGAGMGGGNDEREQTINQLLTEMDGFSGNSGV 389
Query 119 VVLAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALAE 178
+VLA TNR D+LD+AL R GRFDR+V ++RPDV R +I KVH + + VD +A
Sbjct 390 IVLAATNRPDVLDSALLRPGRFDRQVTVDRPDVAGRVQILKVHSRGKAIGKDVDYEKVAR 449
Query 179 R 179
R
Sbjct 450 R 450
> 7294008
Length=793
Score = 226 bits (576), Expect = 2e-59, Method: Compositional matrix adjust.
Identities = 115/185 (62%), Positives = 142/185 (76%), Gaps = 4/185 (2%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
KNP + +GAK+PKGA+L GPPGTGKTLLAKA AGEA VPF ++SGS+F+E+FVGVG S
Sbjct 315 KNPQQYIDLGAKIPKGAMLTGPPGTGKTLLAKATAGEANVPFITVSGSEFLEMFVGVGPS 374
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASRS--TQFANSEREQTLNQLLVEMDGFSPHESV 118
RVR++F ARK AP I++IDEID+VG R T +SE+E TLNQLLVEMDGF+ +V
Sbjct 375 RVRDMFAMARKHAPCILFIDEIDAVGRKRGGKTFGGHSEQENTLNQLLVEMDGFNTTTNV 434
Query 119 VVLAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALAE 178
VVLA TNR D+LD AL R GRFDR++ + PD+K RA IFKVHL L+ S +D N L+
Sbjct 435 VVLAATNRVDILDKALMRPGRFDRQIYVPAPDIKGRASIFKVHLGNLKTS--LDKNELSR 492
Query 179 RMAAL 183
+MAAL
Sbjct 493 KMAAL 497
> At2g30950
Length=695
Score = 224 bits (571), Expect = 8e-59, Method: Compositional matrix adjust.
Identities = 111/181 (61%), Positives = 136/181 (75%), Gaps = 2/181 (1%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
K P F +GAK+PKG LL+GPPGTGKTLLAKA+AGEAGVPFFSISGS+FVE+FVGVGAS
Sbjct 247 KKPERFTAVGAKIPKGVLLIGPPGTGKTLLAKAIAGEAGVPFFSISGSEFVEMFVGVGAS 306
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASRSTQF--ANSEREQTLNQLLVEMDGFSPHESV 118
RVR+LF +A++ AP I+++DEID+VG R T N EREQTLNQLL EMDGF + V
Sbjct 307 RVRDLFKKAKENAPCIVFVDEIDAVGRQRGTGIGGGNDEREQTLNQLLTEMDGFEGNTGV 366
Query 119 VVLAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALAE 178
+V+A TNR D+LD+AL R GRFDR+V ++ PDVK R +I KVH + V +A
Sbjct 367 IVVAATNRADILDSALLRPGRFDRQVSVDVPDVKGRTDILKVHAGNKKFDNDVSLEIIAM 426
Query 179 R 179
R
Sbjct 427 R 427
> At1g06430
Length=685
Score = 224 bits (570), Expect = 1e-58, Method: Compositional matrix adjust.
Identities = 111/181 (61%), Positives = 136/181 (75%), Gaps = 2/181 (1%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
K P F +GA++PKG LLVGPPGTGKTLLAKA+AGEAGVPFFSISGS+FVE+FVGVGAS
Sbjct 240 KKPERFTAVGARIPKGVLLVGPPGTGKTLLAKAIAGEAGVPFFSISGSEFVEMFVGVGAS 299
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASRSTQF--ANSEREQTLNQLLVEMDGFSPHESV 118
RVR+LF +A++ AP I+++DEID+VG R T N EREQTLNQLL EMDGF + V
Sbjct 300 RVRDLFKKAKENAPCIVFVDEIDAVGRQRGTGIGGGNDEREQTLNQLLTEMDGFEGNTGV 359
Query 119 VVLAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALAE 178
+V+A TNR D+LD+AL R GRFDR+V ++ PDVK R +I KVH + V +A
Sbjct 360 IVVAATNRADILDSALLRPGRFDRQVSVDVPDVKGRTDILKVHSGNKKFESGVSLEVIAM 419
Query 179 R 179
R
Sbjct 420 R 420
> At5g15250
Length=687
Score = 219 bits (557), Expect = 3e-57, Method: Compositional matrix adjust.
Identities = 106/181 (58%), Positives = 137/181 (75%), Gaps = 2/181 (1%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
K P F +GAK+PKG LL GPPGTGKTLLAKA+AGEAGVPFFS+SGS+F+E+FVGVGAS
Sbjct 243 KTPEKFSALGAKIPKGVLLTGPPGTGKTLLAKAIAGEAGVPFFSLSGSEFIEMFVGVGAS 302
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASRSTQF--ANSEREQTLNQLLVEMDGFSPHESV 118
R R+LF++A+ +P I++IDEID+VG R T N EREQTLNQ+L EMDGF+ + V
Sbjct 303 RARDLFNKAKANSPCIVFIDEIDAVGRMRGTGIGGGNDEREQTLNQILTEMDGFAGNTGV 362
Query 119 VVLAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALAE 178
+V+A TNR ++LD+AL R GRFDR+V + PD++ R EI KVH + +L V + +A
Sbjct 363 IVIAATNRPEILDSALLRPGRFDRQVSVGLPDIRGREEILKVHSRSKKLDKDVSLSVIAM 422
Query 179 R 179
R
Sbjct 423 R 423
> At1g50250
Length=511
Score = 216 bits (551), Expect = 2e-56, Method: Compositional matrix adjust.
Identities = 105/161 (65%), Positives = 128/161 (79%), Gaps = 2/161 (1%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
KNP + +GAK+PKG LLVGPPGTGKTLLA+AVAGEAGVPFFS + S+FVELFVGVGAS
Sbjct 282 KNPDKYTALGAKIPKGCLLVGPPGTGKTLLARAVAGEAGVPFFSCAASEFVELFVGVGAS 341
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASRSTQF--ANSEREQTLNQLLVEMDGFSPHESV 118
RVR+LF++A+ AP I++IDEID+VG R N EREQT+NQLL EMDGFS + V
Sbjct 342 RVRDLFEKAKSKAPCIVFIDEIDAVGRQRGAGMGGGNDEREQTINQLLTEMDGFSGNSGV 401
Query 119 VVLAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFK 159
+VLA TNR D+LD+AL R GRFDR+V ++RPDV R +I +
Sbjct 402 IVLAATNRPDVLDSALLRPGRFDRQVTVDRPDVAGRVKILQ 442
> CE24356
Length=852
Score = 216 bits (551), Expect = 2e-56, Method: Compositional matrix adjust.
Identities = 108/186 (58%), Positives = 142/186 (76%), Gaps = 6/186 (3%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
KNP ++ +GAK+PKGA+L GPPGTGKTLLAKA AGEA VPF ++SGS+F+E+FVGVG +
Sbjct 313 KNPQQYKDLGAKIPKGAILTGPPGTGKTLLAKATAGEANVPFITVSGSEFLEMFVGVGPA 372
Query 61 RVRELFDEARKAAPSIIWIDEIDSV---GASRSTQFANSEREQTLNQLLVEMDGFSPHE- 116
RVR++F ARK +P I++IDEID+V + +SE+E TLNQLLVEMDGF+ E
Sbjct 373 RVRDMFSMARKNSPCILFIDEIDAVGRKRGGKGGMGGHSEQENTLNQLLVEMDGFTTDES 432
Query 117 SVVVLAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANAL 176
SV+V+A TNR D+LD+AL R GRFDR++ + PD+K RA IF+VHL PLR S +D L
Sbjct 433 SVIVIAATNRVDILDSALLRPGRFDRQIYVPVPDIKGRASIFRVHLGPLRTS--LDKTVL 490
Query 177 AERMAA 182
+ ++AA
Sbjct 491 SRKLAA 496
> Hs4507173
Length=795
Score = 210 bits (534), Expect = 1e-54, Method: Compositional matrix adjust.
Identities = 107/186 (57%), Positives = 138/186 (74%), Gaps = 5/186 (2%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
K+P F ++GAK+PKGALL+GPPG GKTLLAKAVA EA VPF +++G +FVE+ G+GA+
Sbjct 329 KSPERFLQLGAKVPKGALLLGPPGCGKTLLAKAVATEAQVPFLAMAGPEFVEVIGGLGAA 388
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASRSTQ---FANSEREQTLNQLLVEMDGFSPHES 117
RVR LF EAR AP I++IDEID+VG RST F+N+E EQTLNQLLVEMDG +
Sbjct 389 RVRSLFKEARARAPCIVYIDEIDAVGKKRSTTMSGFSNTEEEQTLNQLLVEMDGMGTTDH 448
Query 118 VVVLAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALA 177
V+VLA TNR D+LD AL R GR DR V I+ P ++ER EIF+ HL+ L+L+ + +
Sbjct 449 VIVLASTNRADILDGALMRPGRLDRHVFIDLPTLQERREIFEQHLKSLKLTQ--SSTFYS 506
Query 178 ERMAAL 183
+R+A L
Sbjct 507 QRLAEL 512
> At5g53170
Length=806
Score = 209 bits (531), Expect = 3e-54, Method: Compositional matrix adjust.
Identities = 103/177 (58%), Positives = 133/177 (75%), Gaps = 2/177 (1%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
KNP+ F ++G KLPKG LL G PGTGKTLLAKA+AGEAGVPFF +GS+F E+FVGVGA
Sbjct 382 KNPSKFTRLGGKLPKGILLTGAPGTGKTLLAKAIAGEAGVPFFYRAGSEFEEMFVGVGAR 441
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASRSTQFANSEREQTLNQLLVEMDGFSPHESVVV 120
RVR LF A+K AP II+IDEID+VG++R ++++ TL+QLLVEMDGF +E ++V
Sbjct 442 RVRSLFQAAKKKAPCIIFIDEIDAVGSTRKQWEGHTKK--TLHQLLVEMDGFEQNEGIIV 499
Query 121 LAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALA 177
+A TN D+LD AL R GRFDR +V+ PDV+ R EI +++LQ +S VD A+A
Sbjct 500 MAATNLPDILDPALTRPGRFDRHIVVPSPDVRGREEILELYLQGKPMSEDVDVKAIA 556
> 7290347
Length=819
Score = 208 bits (530), Expect = 4e-54, Method: Compositional matrix adjust.
Identities = 105/189 (55%), Positives = 140/189 (74%), Gaps = 9/189 (4%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
K+P +Q++GAK+P+GALL+GPPG GKTLLAKAVA EA VPF S++GS+F+E+ G+GA+
Sbjct 361 KSPEKYQRLGAKVPRGALLLGPPGCGKTLLAKAVATEAQVPFLSMNGSEFIEMIGGLGAA 420
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASRS-----TQFANSEREQTLNQLLVEMDGFSPH 115
RVR+LF E +K AP II+IDEID++G RS Q ++ E EQTLNQLLVEMDG +
Sbjct 421 RVRDLFKEGKKRAPCIIYIDEIDAIGRQRSGTESMGQGSSGESEQTLNQLLVEMDGMATK 480
Query 116 ESVVVLAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRL-SPRVDAN 174
E V++LA TNR D+LD AL R GRFDR ++I+ P + ER EIF+ HL ++L SP
Sbjct 481 EGVLMLASTNRADILDKALLRPGRFDRHILIDLPTLAERKEIFEKHLSSVKLESP---PT 537
Query 175 ALAERMAAL 183
++R+A L
Sbjct 538 TFSQRLARL 546
> At3g02450
Length=622
Score = 208 bits (529), Expect = 6e-54, Method: Compositional matrix adjust.
Identities = 104/178 (58%), Positives = 137/178 (76%), Gaps = 3/178 (1%)
Query 6 FQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGASRVREL 65
++K+GA+LP+G LLVGPPGTGKTLLA+AVAGEAGVPFFS+S S+FVELFVG GA+R+R+L
Sbjct 359 YKKLGARLPRGVLLVGPPGTGKTLLARAVAGEAGVPFFSVSASEFVELFVGRGAARIRDL 418
Query 66 FDEARKAAPSIIWIDEIDSVGASRSTQFANSEREQTLNQLLVEMDGFSPHESVVVLAGTN 125
F+ ARK +PSII+IDE+D+VG R F N ER+QTLNQLL EMDGF V+V+A TN
Sbjct 419 FNAARKNSPSIIFIDELDAVGGKRGRSF-NDERDQTLNQLLTEMDGFESDTKVIVIAATN 477
Query 126 REDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALAERMAAL 183
R + LD+AL R GRF R+V++ PD + R +I +HL+ + L DA + + +A+L
Sbjct 478 RPEALDSALCRPGRFSRKVLVAEPDQEGRRKILAIHLRDVPLEE--DAFLICDLVASL 533
> Hs7657689
Length=716
Score = 205 bits (522), Expect = 4e-53, Method: Compositional matrix adjust.
Identities = 101/177 (57%), Positives = 124/177 (70%), Gaps = 0/177 (0%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
KNP F +G KLPKG LLVGPPGTGKTLLA+AVAGEA VPF+ SGS+F E+FVGVGAS
Sbjct 302 KNPQKFTILGGKLPKGILLVGPPGTGKTLLARAVAGEADVPFYYASGSEFDEMFVGVGAS 361
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASRSTQFANSEREQTLNQLLVEMDGFSPHESVVV 120
R+R LF EA+ AP +I+IDE+DSVG R + QT+NQLL EMDGF P+E V++
Sbjct 362 RIRNLFREAKANAPCVIFIDELDSVGGKRIESPMHPYSRQTINQLLAEMDGFKPNEGVII 421
Query 121 LAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALA 177
+ TN + LD AL R GRFD +V + RPDVK R EI K +L ++ VD +A
Sbjct 422 IGATNFPEALDNALIRPGRFDMQVTVPRPDVKGRTEILKWYLNKIKFDQSVDPEIIA 478
> Hs21327685
Length=773
Score = 205 bits (521), Expect = 4e-53, Method: Compositional matrix adjust.
Identities = 101/177 (57%), Positives = 124/177 (70%), Gaps = 0/177 (0%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
KNP F +G KLPKG LLVGPPGTGKTLLA+AVAGEA VPF+ SGS+F E+FVGVGAS
Sbjct 359 KNPQKFTILGGKLPKGILLVGPPGTGKTLLARAVAGEADVPFYYASGSEFDEMFVGVGAS 418
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASRSTQFANSEREQTLNQLLVEMDGFSPHESVVV 120
R+R LF EA+ AP +I+IDE+DSVG R + QT+NQLL EMDGF P+E V++
Sbjct 419 RIRNLFREAKANAPCVIFIDELDSVGGKRIESPMHPYSRQTINQLLAEMDGFKPNEGVII 478
Query 121 LAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALA 177
+ TN + LD AL R GRFD +V + RPDVK R EI K +L ++ VD +A
Sbjct 479 IGATNFPEALDNALIRPGRFDMQVTVPRPDVKGRTEILKWYLNKIKFDQSVDPEIIA 535
> Hs21327687
Length=740
Score = 205 bits (521), Expect = 4e-53, Method: Compositional matrix adjust.
Identities = 101/177 (57%), Positives = 124/177 (70%), Gaps = 0/177 (0%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
KNP F +G KLPKG LLVGPPGTGKTLLA+AVAGEA VPF+ SGS+F E+FVGVGAS
Sbjct 359 KNPQKFTILGGKLPKGILLVGPPGTGKTLLARAVAGEADVPFYYASGSEFDEMFVGVGAS 418
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASRSTQFANSEREQTLNQLLVEMDGFSPHESVVV 120
R+R LF EA+ AP +I+IDE+DSVG R + QT+NQLL EMDGF P+E V++
Sbjct 419 RIRNLFREAKANAPCVIFIDELDSVGGKRIESPMHPYSRQTINQLLAEMDGFKPNEGVII 478
Query 121 LAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALA 177
+ TN + LD AL R GRFD +V + RPDVK R EI K +L ++ VD +A
Sbjct 479 IGATNFPEALDNALIRPGRFDMQVTVPRPDVKGRTEILKWYLNKIKFDQSVDPEIIA 535
> At5g58870
Length=806
Score = 204 bits (519), Expect = 9e-53, Method: Compositional matrix adjust.
Identities = 106/177 (59%), Positives = 127/177 (71%), Gaps = 3/177 (1%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
KNP + ++GA+ P+G LLVG PGTGKTLLAKAVAGE+ VPF S S S+FVEL+VG+GAS
Sbjct 349 KNPDRYVRLGARPPRGVLLVGLPGTGKTLLAKAVAGESDVPFISCSASEFVELYVGMGAS 408
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASRSTQF---ANSEREQTLNQLLVEMDGFSPHES 117
RVR+LF A+K APSII+IDEID+V SR +F +N EREQTLNQLL EMDGF +
Sbjct 409 RVRDLFARAKKEAPSIIFIDEIDAVAKSRDGKFRMVSNDEREQTLNQLLTEMDGFDSSSA 468
Query 118 VVVLAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDAN 174
V+VL TNR D+LD AL+R GRFDR V + PD R I KVH+ L D N
Sbjct 469 VIVLGATNRADVLDPALRRPGRFDRVVTVESPDKVGRESILKVHVSKKELPLGDDVN 525
> At3g47060
Length=802
Score = 204 bits (518), Expect = 1e-52, Method: Compositional matrix adjust.
Identities = 105/177 (59%), Positives = 128/177 (72%), Gaps = 3/177 (1%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
+NP + ++GA+ P+G LLVG PGTGKTLLAKAVAGEA VPF S S S+FVEL+VG+GAS
Sbjct 345 RNPEKYVRLGARPPRGVLLVGLPGTGKTLLAKAVAGEAEVPFISCSASEFVELYVGMGAS 404
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASRSTQF---ANSEREQTLNQLLVEMDGFSPHES 117
RVR+LF A+K APSII+IDEID+V SR +F +N EREQTLNQLL EMDGF + +
Sbjct 405 RVRDLFARAKKEAPSIIFIDEIDAVAKSRDGKFRMGSNDEREQTLNQLLTEMDGFDSNSA 464
Query 118 VVVLAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDAN 174
V+VL TNR D+LD AL+R GRFDR V + PD R I +VH+ L D N
Sbjct 465 VIVLGATNRADVLDPALRRPGRFDRVVTVETPDKIGRESILRVHVSKKELPLGDDVN 521
> At2g26140
Length=627
Score = 199 bits (506), Expect = 3e-51, Method: Compositional matrix adjust.
Identities = 96/177 (54%), Positives = 130/177 (73%), Gaps = 1/177 (0%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
++P F ++G KLPKG LLVGPPGTGKT+LA+A+AGEAGVPFFS SGS+F E+FVGVGA
Sbjct 157 RDPKRFTRLGGKLPKGVLLVGPPGTGKTMLARAIAGEAGVPFFSCSGSEFEEMFVGVGAR 216
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASRSTQFANSEREQTLNQLLVEMDGFSPHESVVV 120
RVR+LF A+K +P II+IDEID++G SR+ + + TLNQ+LVE+DGF +E ++V
Sbjct 217 RVRDLFSAAKKCSPCIIFIDEIDAIGGSRNPK-DQQYMKMTLNQMLVELDGFKQNEGIIV 275
Query 121 LAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALA 177
+A TN + LD AL R GRFDR +V+ PDV+ R +I + H+ + + VD +A
Sbjct 276 VAATNFPESLDKALVRPGRFDRHIVVPNPDVEGRRQILESHMSKVLKAEDVDLMIIA 332
> CE03496
Length=676
Score = 199 bits (505), Expect = 3e-51, Method: Compositional matrix adjust.
Identities = 97/178 (54%), Positives = 128/178 (71%), Gaps = 0/178 (0%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
K+P + ++G +LPKG LLVGPPGTGKTLLA+A+AGEA VPFF +GS+F E+ VG GA
Sbjct 221 KDPEKYSRLGGRLPKGVLLVGPPGTGKTLLARAIAGEAQVPFFHTAGSEFDEVLVGQGAR 280
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASRSTQFANSEREQTLNQLLVEMDGFSPHESVVV 120
RVR+LFD+A+ AP II+IDEIDSVG+ R + + QT+NQLL EMDGF+ +E ++V
Sbjct 281 RVRDLFDKAKARAPCIIFIDEIDSVGSKRVSNSIHPYANQTINQLLSEMDGFTRNEGIIV 340
Query 121 LAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALAE 178
+A TNR D LD AL R GRFD RV + +PD+ R +IF +L + S +D LA+
Sbjct 341 IAATNRVDDLDKALLRPGRFDVRVTVPKPDLAGRVDIFNFYLSKIVHSGGIDPKVLAK 398
> At4g23940
Length=946
Score = 195 bits (495), Expect = 5e-50, Method: Compositional matrix adjust.
Identities = 96/190 (50%), Positives = 128/190 (67%), Gaps = 10/190 (5%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
KNP F KMG K P G LL GPPG GKTL+AKA+AGEAGVPF+ ++GS+FVE+ VGVG++
Sbjct 450 KNPDLFDKMGIKPPHGVLLEGPPGCGKTLVAKAIAGEAGVPFYQMAGSEFVEVLVGVGSA 509
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASRSTQF----------ANSEREQTLNQLLVEMD 110
R+R+LF A+ PS+I+IDEID++ R F A ERE TLNQLL+E+D
Sbjct 510 RIRDLFKRAKVNKPSVIFIDEIDALATRRQGIFKENSDQLYNAATQERETTLNQLLIELD 569
Query 111 GFSPHESVVVLAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPR 170
GF + V+ L TNR DLLD AL R GRFDR++ + P+ K R +I K+H +++S
Sbjct 570 GFDTGKGVIFLGATNRRDLLDPALLRPGRFDRKIRVRPPNAKGRLDILKIHASKVKMSDS 629
Query 171 VDANALAERM 180
VD ++ A +
Sbjct 630 VDLSSYASNL 639
> YPR024w
Length=747
Score = 194 bits (494), Expect = 6e-50, Method: Compositional matrix adjust.
Identities = 93/177 (52%), Positives = 130/177 (73%), Gaps = 1/177 (0%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
K+P ++ +G KLPKG LL GPPGTGKTLLA+A AGEAGV FF +SGS+F E++VGVGA
Sbjct 301 KDPTKYESLGGKLPKGVLLTGPPGTGKTLLARATAGEAGVDFFFMSGSEFDEVYVGVGAK 360
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASRSTQFANSEREQTLNQLLVEMDGFSPHESVVV 120
R+R+LF +AR AP+II+IDE+D++G R+ + + +QTLNQLLVE+DGFS +++
Sbjct 361 RIRDLFAQARSRAPAIIFIDELDAIGGKRNPK-DQAYAKQTLNQLLVELDGFSQTSGIII 419
Query 121 LAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALA 177
+ TN + LD AL R GRFD+ V ++ PDV+ RA+I K H++ + L+ VD +A
Sbjct 420 IGATNFPEALDKALTRPGRFDKVVNVDLPDVRGRADILKHHMKKITLADNVDPTIIA 476
> 7291497
Length=736
Score = 189 bits (481), Expect = 2e-48, Method: Compositional matrix adjust.
Identities = 97/177 (54%), Positives = 120/177 (67%), Gaps = 1/177 (0%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
K+P F +G KLPKG LLVGPPGTGKTLLA+AVAGEA VPFF +G +F E+ VG GA
Sbjct 320 KSPEKFSNLGGKLPKGVLLVGPPGTGKTLLARAVAGEAKVPFFHAAGPEFDEVLVGQGAR 379
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASRSTQFANSEREQTLNQLLVEMDGFSPHESVVV 120
RVR+LF A+ AP +I+IDEIDSVGA R+ + QT+NQLL EMDGF + V+V
Sbjct 380 RVRDLFKAAKARAPCVIFIDEIDSVGAKRTNSVLHPYANQTINQLLSEMDGFHQNAGVIV 439
Query 121 LAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALA 177
L TNR D LD AL R GRFD V+++ PD R EI ++L + L +D + LA
Sbjct 440 LGATNRRDDLDQALLRPGRFDVEVMVSTPDFTGRKEILSLYLTKI-LHDEIDLDMLA 495
> SPCC965.04c
Length=709
Score = 186 bits (472), Expect = 2e-47, Method: Compositional matrix adjust.
Identities = 98/181 (54%), Positives = 126/181 (69%), Gaps = 5/181 (2%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
++P F ++G KLP+G LL GPPGTGKT+LA+AVAGEA VPFF +SGS F E++VGVGA
Sbjct 287 RDPTHFTRLGGKLPRGVLLTGPPGTGKTMLARAVAGEANVPFFFMSGSQFDEMYVGVGAK 346
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASRSTQFANSEREQTLNQLLVEMDGFSPHES--- 117
RVRELF ARK APSII+IDE+D++G R+ + A R QTLNQLLV++DGFS +E
Sbjct 347 RVRELFAAARKQAPSIIFIDELDAIGQKRNARDAAHMR-QTLNQLLVDLDGFSKNEDLAH 405
Query 118 -VVVLAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANAL 176
VV + TN + LD AL R GRFDR + + PDV+ R I H + + L VD + +
Sbjct 406 PVVFIGATNFPESLDPALTRPGRFDRHIHVPLPDVRGRLAILLQHTRHVPLGKDVDLSII 465
Query 177 A 177
A
Sbjct 466 A 466
> At3g16290
Length=876
Score = 185 bits (470), Expect = 4e-47, Method: Compositional matrix adjust.
Identities = 95/174 (54%), Positives = 123/174 (70%), Gaps = 2/174 (1%)
Query 6 FQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGASRVREL 65
+++ G K+P G LL GPPG GKTLLAKAVAGEAGV FFSIS S FVE++VGVGASRVR L
Sbjct 435 YRRRGVKIPGGILLCGPPGVGKTLLAKAVAGEAGVNFFSISASQFVEIYVGVGASRVRAL 494
Query 66 FDEARKAAPSIIWIDEIDSVGASRS--TQFANSEREQTLNQLLVEMDGFSPHESVVVLAG 123
+ EAR+ APS+++IDE+D+VG R ER+ TLNQLLV +DGF V+ +A
Sbjct 495 YQEARENAPSVVFIDELDAVGRERGLIKGSGGQERDATLNQLLVSLDGFEGRGEVITIAS 554
Query 124 TNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALA 177
TNR D+LD AL R GRFDR++ I +P + R EI +VH + ++ +D A+A
Sbjct 555 TNRPDILDPALVRPGRFDRKIFIPKPGLIGRMEILQVHARKKPMAEDLDYMAVA 608
> At5g64580
Length=871
Score = 177 bits (449), Expect = 1e-44, Method: Compositional matrix adjust.
Identities = 93/165 (56%), Positives = 114/165 (69%), Gaps = 4/165 (2%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
KN FQ G PKG LL GPPGTGKTLLAKA+AGEAG+PFF+ +G+DFVE+FVGV AS
Sbjct 336 KNDEEFQNKGIYCPKGVLLHGPPGTGKTLLAKAIAGEAGLPFFAANGTDFVEMFVGVAAS 395
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASR---STQFANSEREQTLNQLLVEMDGFSPHES 117
RV++LF +R APSII+IDEID++G+ R +EREQ L Q+L EMDGF S
Sbjct 396 RVKDLFASSRSYAPSIIFIDEIDAIGSKRGGPDIGGGGAEREQGLLQILTEMDGFKVTTS 455
Query 118 -VVVLAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVH 161
V+V+ TNR D+LD AL R GRFD+ + + P R I KVH
Sbjct 456 QVLVIGATNRLDILDPALLRKGRFDKIIRVGLPSKDGRLAILKVH 500
> ECU09g1840
Length=453
Score = 171 bits (432), Expect = 1e-42, Method: Compositional matrix adjust.
Identities = 83/185 (44%), Positives = 122/185 (65%), Gaps = 2/185 (1%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
K+P F+ +G PKG LL GPPGTGKTLLA+AVA F +SGS+ V+ ++G G+
Sbjct 217 KHPELFENLGIAQPKGVLLYGPPGTGKTLLARAVAHHTQCKFIRVSGSELVQKYIGEGSR 276
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASR--STQFANSEREQTLNQLLVEMDGFSPHESV 118
VRELF AR+ APSII++DEIDS+G++R S + ++SE ++T+ +LL ++DGF H ++
Sbjct 277 LVRELFIMAREHAPSIIFMDEIDSIGSTRGDSNKGSDSEVQRTMLELLNQLDGFESHNNI 336
Query 119 VVLAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALAE 178
V+ TNR D+LD AL R GR DR++ P+ R EI K+H + + L+ +D +A
Sbjct 337 KVIMATNRIDILDPALLRTGRIDRKIEFPPPNESARLEILKIHSRKMNLTKGIDLETIAS 396
Query 179 RMAAL 183
+M
Sbjct 397 KMVGC 401
> Hs22047546
Length=611
Score = 168 bits (425), Expect = 6e-42, Method: Compositional matrix adjust.
Identities = 92/187 (49%), Positives = 123/187 (65%), Gaps = 15/187 (8%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
K+ A +Q +GAK+PKGA+L GPPGTGKTLLAKA AG+A VPF ++SGS+F+E+FV +
Sbjct 280 KDVAQYQDLGAKIPKGAILTGPPGTGKTLLAKATAGKANVPFITVSGSEFLEMFVE--SE 337
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASRSTQFAN----SEREQTLNQLLVEMDGFSPHE 116
L A+ S + R T N +E+E TLNQLLVEMDGF+
Sbjct 338 TYLPLLGRMPLASSSNV---------VGRKTGKGNFGGQNEQEDTLNQLLVEMDGFNTTT 388
Query 117 SVVVLAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANAL 176
+VV+LA TN+ D+LD AL R GRFDR++ I PD+K R IFKVHL+PL+L ++ L
Sbjct 389 NVVILASTNQPDILDPALLRPGRFDRQIFIGLPDIKGRGSIFKVHLRPLKLDSTLEKEKL 448
Query 177 AERMAAL 183
A ++A+L
Sbjct 449 ARKLASL 455
> CE28467
Length=411
Score = 166 bits (420), Expect = 2e-41, Method: Compositional matrix adjust.
Identities = 85/182 (46%), Positives = 123/182 (67%), Gaps = 2/182 (1%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
K+P F +G PKG LL GPPGTGKTLLA+AVA F +SGS+ V+ F+G GA
Sbjct 175 KHPELFDALGIAQPKGVLLYGPPGTGKTLLARAVAHHTECTFIRVSGSELVQKFIGEGAR 234
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASR--STQFANSEREQTLNQLLVEMDGFSPHESV 118
VRELF AR+ APSII++DEIDS+G+SR ++ +SE ++T+ +LL ++DGF +++
Sbjct 235 MVRELFVMAREHAPSIIFMDEIDSIGSSRVEGSRGGDSEVQRTMLELLNQLDGFEATKNI 294
Query 119 VVLAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALAE 178
V+ TNR D+LD+AL R GR DR++ PD K RA+I K+H + + L + + +AE
Sbjct 295 KVIMATNRIDILDSALLRPGRIDRKIEFPAPDEKARAQILKIHSRKMNLMRGIRMDKIAE 354
Query 179 RM 180
++
Sbjct 355 QI 356
> CE22219
Length=416
Score = 164 bits (415), Expect = 9e-41, Method: Compositional matrix adjust.
Identities = 85/182 (46%), Positives = 121/182 (66%), Gaps = 2/182 (1%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
K+P F +G PKG LL GPPGTGKTLLA+AVA F +SGS+ V+ F+G GA
Sbjct 180 KHPELFDALGIAQPKGVLLFGPPGTGKTLLARAVAHHTECTFIRVSGSELVQKFIGEGAR 239
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASR--STQFANSEREQTLNQLLVEMDGFSPHESV 118
VRELF AR+ APSII++DEIDS+G+SR + +SE ++T+ +LL ++DGF +++
Sbjct 240 MVRELFVMAREHAPSIIFMDEIDSIGSSRVEGSSGGDSEVQRTMLELLNQLDGFEATKNI 299
Query 119 VVLAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALAE 178
V+ TNR D+LD AL R GR DR++ PD K RA+I K+H + + L ++ +AE
Sbjct 300 KVIMATNRIDILDPALLRPGRIDRKIEFPAPDEKARADILKIHSRKMNLMRGINMAKIAE 359
Query 179 RM 180
++
Sbjct 360 QI 361
> At5g19990
Length=405
Score = 163 bits (412), Expect = 2e-40, Method: Compositional matrix adjust.
Identities = 82/183 (44%), Positives = 119/183 (65%), Gaps = 3/183 (1%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
K+P F+ +G PKG LL GPPGTGKTLLA+AVA F +SGS+ V+ ++G G+
Sbjct 168 KHPELFESLGIAQPKGVLLYGPPGTGKTLLARAVAHHTDCTFIRVSGSELVQKYIGEGSR 227
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASR---STQFANSEREQTLNQLLVEMDGFSPHES 117
VRELF AR+ APSII++DEIDS+G++R + +SE ++T+ +LL ++DGF
Sbjct 228 MVRELFVMAREHAPSIIFMDEIDSIGSARMESGSGNGDSEVQRTMLELLNQLDGFEASNK 287
Query 118 VVVLAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALA 177
+ VL TNR D+LD AL R GR DR++ P+ + R +I K+H + + L +D +A
Sbjct 288 IKVLMATNRIDILDQALLRPGRIDRKIEFPNPNEESRFDILKIHSRKMNLMRGIDLKKIA 347
Query 178 ERM 180
E+M
Sbjct 348 EKM 350
> At5g20000
Length=419
Score = 162 bits (411), Expect = 2e-40, Method: Compositional matrix adjust.
Identities = 82/183 (44%), Positives = 119/183 (65%), Gaps = 3/183 (1%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
K+P F+ +G PKG LL GPPGTGKTLLA+AVA F +SGS+ V+ ++G G+
Sbjct 182 KHPELFESLGIAQPKGVLLYGPPGTGKTLLARAVAHHTDCTFIRVSGSELVQKYIGEGSR 241
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASR---STQFANSEREQTLNQLLVEMDGFSPHES 117
VRELF AR+ APSII++DEIDS+G++R + +SE ++T+ +LL ++DGF
Sbjct 242 MVRELFVMAREHAPSIIFMDEIDSIGSARMESGSGNGDSEVQRTMLELLNQLDGFEASNK 301
Query 118 VVVLAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALA 177
+ VL TNR D+LD AL R GR DR++ P+ + R +I K+H + + L +D +A
Sbjct 302 IKVLMATNRIDILDQALLRPGRIDRKIEFPNPNEESRFDILKIHSRKMNLMRGIDLKKIA 361
Query 178 ERM 180
E+M
Sbjct 362 EKM 364
> YGL048c
Length=405
Score = 161 bits (408), Expect = 5e-40, Method: Compositional matrix adjust.
Identities = 79/182 (43%), Positives = 122/182 (67%), Gaps = 2/182 (1%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
K+P F+ +G PKG +L GPPGTGKTLLA+AVA F +SG++ V+ ++G G+
Sbjct 169 KHPELFESLGIAQPKGVILYGPPGTGKTLLARAVAHHTDCKFIRVSGAELVQKYIGEGSR 228
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASR--STQFANSEREQTLNQLLVEMDGFSPHESV 118
VRELF AR+ APSII++DEIDS+G++R + +SE ++T+ +LL ++DGF +++
Sbjct 229 MVRELFVMAREHAPSIIFMDEIDSIGSTRVEGSGGGDSEVQRTMLELLNQLDGFETSKNI 288
Query 119 VVLAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALAE 178
++ TNR D+LD AL R GR DR++ P V RAEI ++H + + L+ ++ +AE
Sbjct 289 KIIMATNRLDILDPALLRPGRIDRKIEFPPPSVAARAEILRIHSRKMNLTRGINLRKVAE 348
Query 179 RM 180
+M
Sbjct 349 KM 350
> Hs14771027
Length=406
Score = 161 bits (408), Expect = 5e-40, Method: Compositional matrix adjust.
Identities = 83/182 (45%), Positives = 121/182 (66%), Gaps = 2/182 (1%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
K+P F+ +G PKG LL GPPGTGKTLLA+AVA F +SGS+ V+ F+G GA
Sbjct 170 KHPELFEALGIAQPKGVLLYGPPGTGKTLLARAVAHHTDCTFIRVSGSELVQKFIGEGAR 229
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASR--STQFANSEREQTLNQLLVEMDGFSPHESV 118
VRELF AR+ APSII++DEIDS+G+SR +SE ++T+ +LL ++DGF +++
Sbjct 230 MVRELFVMAREHAPSIIFMDEIDSIGSSRLEGGSGGDSEVQRTMLELLNQLDGFEATKNI 289
Query 119 VVLAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALAE 178
V+ TNR D+LD+AL R GR DR++ P+ + R +I K+H + + L+ ++ +AE
Sbjct 290 KVIMATNRIDILDSALLRPGRIDRKIEFPPPNEEARLDILKIHSRKMNLTRGINLRKIAE 349
Query 179 RM 180
M
Sbjct 350 LM 351
> 7301963
Length=399
Score = 161 bits (407), Expect = 7e-40, Method: Compositional matrix adjust.
Identities = 82/181 (45%), Positives = 120/181 (66%), Gaps = 1/181 (0%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
K+P F +G PKG LL GPPGTGKTLLA+AVA F +SGS+ V+ F+G G+
Sbjct 164 KHPELFDALGITQPKGVLLYGPPGTGKTLLARAVAHHTECTFIRVSGSELVQKFIGEGSR 223
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASR-STQFANSEREQTLNQLLVEMDGFSPHESVV 119
VRELF AR+ APSII++DEIDS+G++R T +SE ++T+ +LL ++DGF +++
Sbjct 224 MVRELFVMAREHAPSIIFMDEIDSIGSARLETGTGDSEVQRTMLELLNQLDGFEATKNIK 283
Query 120 VLAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALAER 179
V+ TNR D+LD AL R GR DR++ P+ + R +I K+H + + L+ ++ +AE
Sbjct 284 VIMATNRIDVLDQALLRPGRIDRKIEFPPPNEEARLDILKIHSRKMNLTRGINLRKIAEE 343
Query 180 M 180
M
Sbjct 344 M 344
> 7295522
Length=405
Score = 160 bits (405), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 83/182 (45%), Positives = 120/182 (65%), Gaps = 2/182 (1%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
K+P F +G PKG LL GPPGTGKTLLA+AVA F +SGS+ V+ F+G G+
Sbjct 169 KHPELFDALGIAQPKGVLLYGPPGTGKTLLARAVAHHTECTFIRVSGSELVQKFIGEGSR 228
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASR--STQFANSEREQTLNQLLVEMDGFSPHESV 118
VRELF AR+ APSII++DEIDS+G+SR S +SE ++T+ +LL ++DGF +++
Sbjct 229 MVRELFVMAREHAPSIIFMDEIDSIGSSRIESGSGGDSEVQRTMLELLNQLDGFEATKNI 288
Query 119 VVLAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALAE 178
V+ TNR D+LD AL R GR DR++ P+ + R +I K+H + + L+ ++ +AE
Sbjct 289 KVIMATNRIDILDPALLRPGRIDRKIEFPPPNEEARLDILKIHSRKMNLTRGINLRKIAE 348
Query 179 RM 180
M
Sbjct 349 LM 350
> Hs4506213
Length=406
Score = 159 bits (402), Expect = 3e-39, Method: Compositional matrix adjust.
Identities = 82/182 (45%), Positives = 120/182 (65%), Gaps = 2/182 (1%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
K+P F+ +G PKG LL GPPGTGKTLLA+AVA F +SGS+ V+ F+G GA
Sbjct 170 KHPELFEALGIAQPKGVLLYGPPGTGKTLLARAVAHHTDCTFIRVSGSELVQKFIGEGAR 229
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASR--STQFANSEREQTLNQLLVEMDGFSPHESV 118
VRELF AR+ APSII++DEIDS+G+SR +SE ++ + +LL ++DGF +++
Sbjct 230 MVRELFVMAREHAPSIIFMDEIDSIGSSRLEGGSGGSSEVQRQMLELLNQLDGFEATKNI 289
Query 119 VVLAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALAE 178
V+ TNR D+LD+AL R GR DR++ P+ + R +I K+H + + L+ ++ +AE
Sbjct 290 KVIMATNRIDMLDSALLRPGRIDRKIEFPPPNEEARLDILKIHSRKMNLTRGINLRKIAE 349
Query 179 RM 180
M
Sbjct 350 LM 351
> At3g05530
Length=424
Score = 156 bits (395), Expect = 2e-38, Method: Compositional matrix adjust.
Identities = 76/174 (43%), Positives = 113/174 (64%), Gaps = 2/174 (1%)
Query 6 FQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGASRVREL 65
F+K+G + PKG LL GPPGTGKTL+A+A A + F ++G V++F+G GA VR+
Sbjct 197 FEKLGVRPPKGVLLYGPPGTGKTLMARACAAQTNATFLKLAGPQLVQMFIGDGAKLVRDA 256
Query 66 FDEARKAAPSIIWIDEIDSVGASRSTQFANSERE--QTLNQLLVEMDGFSPHESVVVLAG 123
F A++ AP II+IDEID++G R + +RE +T+ +LL ++DGFS E + V+A
Sbjct 257 FQLAKEKAPCIIFIDEIDAIGTKRFDSEVSGDREVQRTMLELLNQLDGFSSDERIKVIAA 316
Query 124 TNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALA 177
TNR D+LD AL R+GR DR++ P + RA I ++H + + + P V+ LA
Sbjct 317 TNRADILDPALMRSGRLDRKIEFPHPTEEARARILQIHSRKMNVHPDVNFEELA 370
> Hs21361144
Length=439
Score = 156 bits (395), Expect = 2e-38, Method: Compositional matrix adjust.
Identities = 76/174 (43%), Positives = 116/174 (66%), Gaps = 2/174 (1%)
Query 6 FQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGASRVREL 65
F+ +G + PKG L+ GPPGTGKTLLA+A A + F ++G V++F+G GA VR+
Sbjct 212 FENLGIQPPKGVLMYGPPGTGKTLLARACAAQTKATFLKLAGPQLVQMFIGDGAKLVRDA 271
Query 66 FDEARKAAPSIIWIDEIDSVGASR--STQFANSEREQTLNQLLVEMDGFSPHESVVVLAG 123
F A++ APSII+IDE+D++G R S + + E ++T+ +LL ++DGF P+ V V+A
Sbjct 272 FALAKEKAPSIIFIDELDAIGTKRFDSEKAGDREVQRTMLELLNQLDGFQPNTQVKVIAA 331
Query 124 TNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALA 177
TNR D+LD AL R+GR DR++ P+ + RA I ++H + + +SP V+ LA
Sbjct 332 TNRVDILDPALLRSGRLDRKIEFPMPNEEARARIMQIHSRKMNVSPDVNYEELA 385
> HsM4506211
Length=404
Score = 156 bits (395), Expect = 2e-38, Method: Compositional matrix adjust.
Identities = 76/174 (43%), Positives = 116/174 (66%), Gaps = 2/174 (1%)
Query 6 FQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGASRVREL 65
F+ +G + PKG L+ GPPGTGKTLLA+A A + F ++G V++F+G GA VR+
Sbjct 177 FENLGIQPPKGVLMYGPPGTGKTLLARACAAQTKATFLKLAGPQLVQMFIGDGAKLVRDA 236
Query 66 FDEARKAAPSIIWIDEIDSVGASR--STQFANSEREQTLNQLLVEMDGFSPHESVVVLAG 123
F A++ APSII+IDE+D++G R S + + E ++T+ +LL ++DGF P+ V V+A
Sbjct 237 FALAKEKAPSIIFIDELDAIGTKRFDSEKAGDREVQRTMLELLNQLDGFQPNTQVKVIAA 296
Query 124 TNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALA 177
TNR D+LD AL R+GR DR++ P+ + RA I ++H + + +SP V+ LA
Sbjct 297 TNRVDILDPALLRSGRLDRKIEFPMPNEEARARIMQIHSRKMNVSPDVNYEELA 350
> YDL126c
Length=835
Score = 155 bits (393), Expect = 3e-38, Method: Compositional matrix adjust.
Identities = 81/177 (45%), Positives = 113/177 (63%), Gaps = 1/177 (0%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
++P F+ +G K P+G L+ GPPGTGKTL+A+AVA E G FF I+G + + G S
Sbjct 235 RHPQLFKAIGIKPPRGVLMYGPPGTGKTLMARAVANETGAFFFLINGPEVMSKMAGESES 294
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASRSTQFANSEREQTLNQLLVEMDGFSPHESVVV 120
+R+ F+EA K AP+II+IDEIDS+ R ER + ++QLL MDG +VVV
Sbjct 295 NLRKAFEEAEKNAPAIIFIDEIDSIAPKRDKTNGEVER-RVVSQLLTLMDGMKARSNVVV 353
Query 121 LAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALA 177
+A TNR + +D AL+R GRFDR V I PD R E+ ++H + ++L+ VD ALA
Sbjct 354 IAATNRPNSIDPALRRFGRFDREVDIGIPDATGRLEVLRIHTKNMKLADDVDLEALA 410
Score = 140 bits (352), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 70/179 (39%), Positives = 106/179 (59%), Gaps = 2/179 (1%)
Query 2 NPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGASR 61
+P + K G KG L GPPGTGKTLLAKAVA E F S+ G + + ++ G S
Sbjct 509 HPDQYTKFGLSPSKGVLFYGPPGTGKTLLAKAVATEVSANFISVKGPELLSMWYGESESN 568
Query 62 VRELFDEARKAAPSIIWIDEIDSVGASRSTQF--ANSEREQTLNQLLVEMDGFSPHESVV 119
+R++FD+AR AAP+++++DE+DS+ +R A ++ +NQLL EMDG + ++V
Sbjct 569 IRDIFDKARAAAPTVVFLDELDSIAKARGGSLGDAGGASDRVVNQLLTEMDGMNAKKNVF 628
Query 120 VLAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALAE 178
V+ TNR D +D A+ R GR D+ + + PD R I L+ L P ++ A+A+
Sbjct 629 VIGATNRPDQIDPAILRPGRLDQLIYVPLPDENARLSILNAQLRKTPLEPGLELTAIAK 687
> Hs6005942
Length=806
Score = 153 bits (387), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 79/177 (44%), Positives = 112/177 (63%), Gaps = 1/177 (0%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
++PA F+ +G K P+G LL GPPGTGKTL+A+AVA E G FF I+G + + G S
Sbjct 225 RHPALFKAIGVKPPRGILLYGPPGTGKTLIARAVANETGAFFFLINGPEIMSKLAGESES 284
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASRSTQFANSEREQTLNQLLVEMDGFSPHESVVV 120
+R+ F+EA K AP+II+IDE+D++ R ER + ++QLL MDG V+V
Sbjct 285 NLRKAFEEAEKNAPAIIFIDELDAIAPKREKTHGEVER-RIVSQLLTLMDGLKQRAHVIV 343
Query 121 LAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALA 177
+A TNR + +D AL+R GRFDR V I PD R EI ++H + ++L+ VD +A
Sbjct 344 MAATNRPNSIDPALRRFGRFDREVDIGIPDATGRLEILQIHTKNMKLADDVDLEQVA 400
Score = 141 bits (356), Expect = 7e-34, Method: Compositional matrix adjust.
Identities = 72/180 (40%), Positives = 108/180 (60%), Gaps = 2/180 (1%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
++P F K G KG L GPPG GKTLLAKA+A E F SI G + + ++ G +
Sbjct 498 EHPDKFLKFGMTPSKGVLFYGPPGCGKTLLAKAIANECQANFISIKGPELLTMWFGESEA 557
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASRSTQFANSER--EQTLNQLLVEMDGFSPHESV 118
VRE+FD+AR+AAP +++ DE+DS+ +R + ++ +NQ+L EMDG S ++V
Sbjct 558 NVREIFDKARQAAPCVLFFDELDSIAKARGGNIGDGGGAADRVINQILTEMDGMSTKKNV 617
Query 119 VVLAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALAE 178
++ TNR D++D A+ R GR D+ + I PD K R I K +L+ ++ VD LA+
Sbjct 618 FIIGATNRPDIIDPAILRPGRLDQLIYIPLPDEKSRVAILKANLRKSPVAKDVDLEFLAK 677
> Hs4506215
Length=389
Score = 152 bits (384), Expect = 3e-37, Method: Compositional matrix adjust.
Identities = 76/179 (42%), Positives = 111/179 (62%), Gaps = 2/179 (1%)
Query 2 NPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGASR 61
NP FQ++G PKG LL GPPGTGKTLLA+AVA + F + S V+ ++G A
Sbjct 155 NPELFQRVGIIPPKGCLLYGPPGTGKTLLARAVASQLDCNFLKVVSSSIVDKYIGESARL 214
Query 62 VRELFDEARKAAPSIIWIDEIDSVGASRSTQFANSERE--QTLNQLLVEMDGFSPHESVV 119
+RE+F+ AR P II++DEID++G R ++ +++RE +TL +LL +MDGF V
Sbjct 215 IREMFNYARDHQPCIIFMDEIDAIGGRRFSEGTSADREIQRTLMELLNQMDGFDTLHRVK 274
Query 120 VLAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALAE 178
+ TNR D LD AL R GR DR++ I+ P+ + R +I K+H P+ +D A+ +
Sbjct 275 MTMATNRPDTLDPALLRPGRLDRKIHIDLPNEQARLDILKIHAGPITKHGEIDYEAIVK 333
> At3g53230
Length=815
Score = 152 bits (384), Expect = 4e-37, Method: Compositional matrix adjust.
Identities = 77/172 (44%), Positives = 108/172 (62%), Gaps = 1/172 (0%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
++P F+ +G K PKG LL GPPG+GKTL+A+AVA E G FF I+G + + G S
Sbjct 229 RHPQLFKSIGVKPPKGILLYGPPGSGKTLIARAVANETGAFFFCINGPEIMSKLAGESES 288
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASRSTQFANSEREQTLNQLLVEMDGFSPHESVVV 120
+R+ F+EA K APSII+IDEIDS+ R ER + ++QLL MDG V+V
Sbjct 289 NLRKAFEEAEKNAPSIIFIDEIDSIAPKREKTHGEVER-RIVSQLLTLMDGLKSRAHVIV 347
Query 121 LAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVD 172
+ TNR + +D AL+R GRFDR + I PD R E+ ++H + ++L+ VD
Sbjct 348 MGATNRPNSIDPALRRFGRFDREIDIGVPDEIGRLEVLRIHTKNMKLAEDVD 399
Score = 148 bits (374), Expect = 5e-36, Method: Compositional matrix adjust.
Identities = 75/180 (41%), Positives = 110/180 (61%), Gaps = 2/180 (1%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
++P F+K G KG L GPPG GKTLLAKA+A E F SI G + + ++ G +
Sbjct 502 EHPEKFEKFGMSPSKGVLFYGPPGCGKTLLAKAIANECQANFISIKGPELLTMWFGESEA 561
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASRSTQF--ANSEREQTLNQLLVEMDGFSPHESV 118
VRE+FD+AR++AP +++ DE+DS+ R A ++ LNQLL EMDG + ++V
Sbjct 562 NVREIFDKARQSAPCVLFFDELDSIATQRGNSVGDAGGAADRVLNQLLTEMDGMNAKKTV 621
Query 119 VVLAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALAE 178
++ TNR D++D AL R GR D+ + I PD + R +IFK L+ ++ VD ALA+
Sbjct 622 FIIGATNRPDIIDPALLRPGRLDQLIYIPLPDEESRYQIFKSCLRKSPVAKDVDLRALAK 681
> ECU01g1230
Length=780
Score = 152 bits (383), Expect = 5e-37, Method: Compositional matrix adjust.
Identities = 82/177 (46%), Positives = 106/177 (59%), Gaps = 1/177 (0%)
Query 3 PAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGASRV 62
P F K G KG L GPPG GKTLLAKAVA E F SI G + + ++VG S +
Sbjct 504 PEKFIKFGMTPAKGVLFYGPPGCGKTLLAKAVATECKANFISIKGPELLSMWVGESESNI 563
Query 63 RELFDEARKAAPSIIWIDEIDSVGASRSTQFANS-EREQTLNQLLVEMDGFSPHESVVVL 121
R+LF AR AAP +++ DEIDS+ +RS +S ++ LNQLL EMDG + ++V V+
Sbjct 564 RDLFARARGAAPCVLFFDEIDSIAKARSGNDGSSGATDRMLNQLLSEMDGINQKKNVFVI 623
Query 122 AGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALAE 178
TNR D LD+AL R GR D+ V I PD+ R I + L+ LSP +D LAE
Sbjct 624 GATNRPDQLDSALMRPGRLDQLVYIPLPDLDSRVSILQATLKKTPLSPEIDLRQLAE 680
Score = 147 bits (371), Expect = 1e-35, Method: Compositional matrix adjust.
Identities = 75/175 (42%), Positives = 109/175 (62%), Gaps = 1/175 (0%)
Query 6 FQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGASRVREL 65
+ K+G K PKG LL GPPGTGKTL+A+A+A E G F I+G + + G S +R+
Sbjct 234 YSKIGVKPPKGILLYGPPGTGKTLIARAIANETGAFLFLINGPEIMSKMAGESESNLRKA 293
Query 66 FDEARKAAPSIIWIDEIDSVGASRSTQFANSEREQTLNQLLVEMDGFSPHESVVVLAGTN 125
F+EA K +P+II+IDEID++ R ER + ++QLL MDG +V+VL TN
Sbjct 294 FEEAEKNSPAIIFIDEIDALAPKREKSQGEVER-RIVSQLLTLMDGMKARSNVIVLGATN 352
Query 126 REDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVDANALAERM 180
R + +D AL+R GRFDR + I PD R EI ++H + +++S VD A+ + +
Sbjct 353 RPNSIDPALRRYGRFDREIEIGVPDETGRLEILRIHTKNMKMSEDVDLVAINKEL 407
> At3g09840
Length=809
Score = 152 bits (383), Expect = 5e-37, Method: Compositional matrix adjust.
Identities = 77/172 (44%), Positives = 108/172 (62%), Gaps = 1/172 (0%)
Query 1 KNPAAFQKMGAKLPKGALLVGPPGTGKTLLAKAVAGEAGVPFFSISGSDFVELFVGVGAS 60
++P F+ +G K PKG LL GPPG+GKTL+A+AVA E G FF I+G + + G S
Sbjct 228 RHPQLFKSIGVKPPKGILLYGPPGSGKTLIARAVANETGAFFFCINGPEIMSKLAGESES 287
Query 61 RVRELFDEARKAAPSIIWIDEIDSVGASRSTQFANSEREQTLNQLLVEMDGFSPHESVVV 120
+R+ F+EA K APSII+IDEIDS+ R ER + ++QLL MDG V+V
Sbjct 288 NLRKAFEEAEKNAPSIIFIDEIDSIAPKREKTNGEVER-RIVSQLLTLMDGLKSRAHVIV 346
Query 121 LAGTNREDLLDAALKRAGRFDRRVVINRPDVKERAEIFKVHLQPLRLSPRVD 172
+ TNR + +D AL+R GRFDR + I PD R E+ ++H + ++L+ VD
Sbjct 347 MGATNRPNSIDPALRRFGRFDREIDIGVPDEIGRLEVLRIHTKNMKLAEDVD 398
Lambda K H
0.318 0.135 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2914594326
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40