bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1200_orf3
Length=161
Score E
Sequences producing significant alignments: (Bits) Value
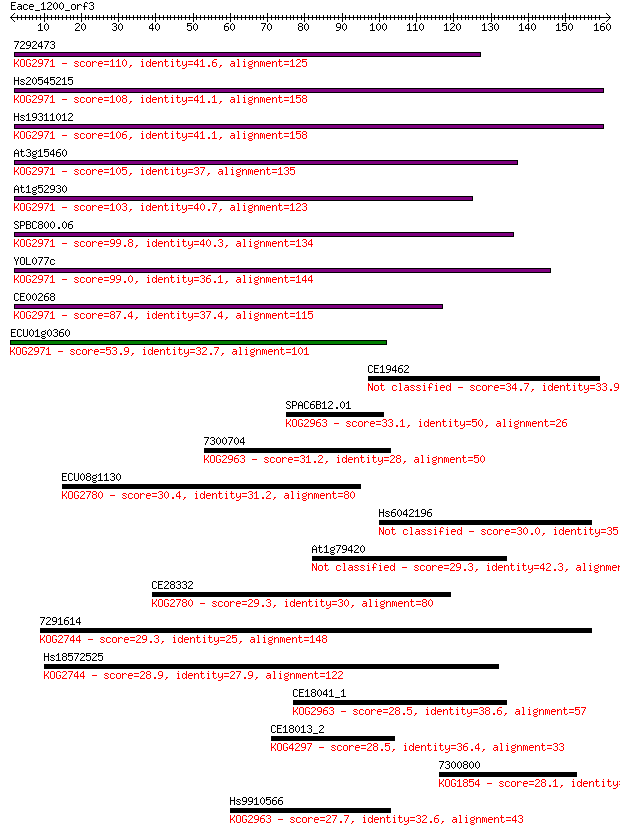
7292473 110 1e-24
Hs20545215 108 6e-24
Hs19311012 106 2e-23
At3g15460 105 4e-23
At1g52930 103 1e-22
SPBC800.06 99.8 2e-21
YOL077c 99.0 4e-21
CE00268 87.4 1e-17
ECU01g0360 53.9 1e-07
CE19462 34.7 0.085
SPAC6B12.01 33.1 0.22
7300704 31.2 1.0
ECU08g1130 30.4 1.5
Hs6042196 30.0 1.8
At1g79420 29.3 3.2
CE28332 29.3 3.5
7291614 29.3 3.7
Hs18572525 28.9 5.0
CE18041_1 28.5 5.7
CE18013_2 28.5 6.3
7300800 28.1 6.8
Hs9910566 27.7 10.0
> 7292473
Length=356
Score = 110 bits (274), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 52/125 (41%), Positives = 76/125 (60%), Gaps = 18/125 (14%)
Query 2 FGVEFDREPHFRVLKELFMQVFGTPRNHPRSKPFFDHAMSFFVTDNRIWFRHYQIAPLVM 61
F +FD PH ++LKELF+Q + P +HP+S+PF DH +F D RIWFR++QI +
Sbjct 157 FDSKFDELPHLKLLKELFVQTYSVPNHHPKSQPFVDHVFTFTYLDKRIWFRNFQI---LS 213
Query 62 GEGGDANNPHGQTFIEIGPRFVLDPVKILEGSFCGKTLWRNAEFVRTRNLMRPRRLREAL 121
+GG E+GPR+V++PVKI +GSF GKT+W N ++V P + R+ L
Sbjct 214 EDGG---------LSEVGPRYVMNPVKIFDGSFTGKTIWENPDYV------SPSKQRQVL 258
Query 122 DFARR 126
A +
Sbjct 259 KKAAK 263
> Hs20545215
Length=353
Score = 108 bits (269), Expect = 6e-24, Method: Compositional matrix adjust.
Identities = 65/161 (40%), Positives = 93/161 (57%), Gaps = 20/161 (12%)
Query 2 FGVEFDREPHFRVLKELFMQVFGTPRNHPRSKPFFDHAMSFFVTDNRIWFRHYQIAPLVM 61
F FD PH+ +LKEL +Q+F TPR HP+S+PF DH +F + DNRIWFR++QI
Sbjct 168 FDPAFDELPHYALLKELLIQIFSTPRYHPKSQPFVDHVFTFTILDNRIWFRNFQII---- 223
Query 62 GEGGDANNPHGQTFIEIGPRFVLDPVKILEGSFCGKTLWRNAEFVRTRNLMRPRRLREAL 121
DA +EIGPRFVL+ +KI +GSF G TL+ N + ++ N+ R R +R
Sbjct 224 --EEDA------ALVEIGPRFVLNLIKIFQGSFGGPTLYENPHY-QSPNMHR-RVIRSIT 273
Query 122 DFARRQGQKEKRRLYLESMLESEPRNKL---ATEEVFGTDA 159
R+ Q+ K ++ + + EP+ L T +VF T A
Sbjct 274 AAKYREKQQVKD---VQKLRKKEPKTLLPHDPTADVFVTPA 311
> Hs19311012
Length=354
Score = 106 bits (265), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 65/161 (40%), Positives = 92/161 (57%), Gaps = 19/161 (11%)
Query 2 FGVEFDREPHFRVLKELFMQVFGTPRNHPRSKPFFDHAMSFFVTDNRIWFRHYQIAPLVM 61
F FD PH+ +LKEL +Q+F TPR HP+S+PF DH +F + DNRIWFR++QI
Sbjct 168 FDPAFDELPHYALLKELLIQIFSTPRYHPKSQPFVDHVFTFTILDNRIWFRNFQII---- 223
Query 62 GEGGDANNPHGQTFIEIGPRFVLDPVKILEGSFCGKTLWRNAEFVRTRNLMRPRRLREAL 121
DA +EIGPRFVL+ +KI +GSF G TL+ N + ++ N+ R R +R
Sbjct 224 --EEDA------ALVEIGPRFVLNLIKIFQGSFGGPTLYENPHY-QSPNMHR-RVIRSIT 273
Query 122 DFARRQGQKEKRRLYLESMLESEPRNKL---ATEEVFGTDA 159
R+ Q+ K + + + EP+ L T +VF T A
Sbjct 274 AAKYREKQQVKD--VPQKLRKKEPKTLLPHDPTADVFVTPA 312
> At3g15460
Length=315
Score = 105 bits (262), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 50/136 (36%), Positives = 79/136 (58%), Gaps = 1/136 (0%)
Query 2 FGVEFDREPHFRVLKELFMQVFGTPRNHPRSKPFFDHAMSFFVTDNRIWFRHYQIA-PLV 60
F F+ + H+++LKE+ Q+FG P H +SKP+ DH F + D+ IWFR+YQI+ P
Sbjct 162 FSSNFENDAHWKLLKEMLTQIFGIPEGHRKSKPYHDHVFVFSIVDDHIWFRNYQISVPHN 221
Query 61 MGEGGDANNPHGQTFIEIGPRFVLDPVKILEGSFCGKTLWRNAEFVRTRNLMRPRRLREA 120
+ + T IE+GPRF L+P+KI GSF G TL+ N +V + + +A
Sbjct 222 ESDKIARGDLDKMTLIEVGPRFCLNPIKIFGGSFGGTTLYENPFYVSPNQIRALEKRNKA 281
Query 121 LDFARRQGQKEKRRLY 136
FA++ K +R+++
Sbjct 282 GKFAKKIKAKTRRKMH 297
> At1g52930
Length=320
Score = 103 bits (258), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 50/124 (40%), Positives = 71/124 (57%), Gaps = 1/124 (0%)
Query 2 FGVEFDREPHFRVLKELFMQVFGTPRNHPRSKPFFDHAMSFFVTDNRIWFRHYQIA-PLV 60
F FD++ H+++LKE+ QVFG P+ H +SKP+ DH F + D IWFR+YQI+ P
Sbjct 166 FSSNFDKDAHWKLLKEMLTQVFGIPKEHRKSKPYHDHVFVFSIVDEHIWFRNYQISVPHN 225
Query 61 MGEGGDANNPHGQTFIEIGPRFVLDPVKILEGSFCGKTLWRNAEFVRTRNLMRPRRLREA 120
+ T IE+GPRF L+P+KI GSF G TL+ N +V + + +A
Sbjct 226 ESDKIAKGGLDKMTLIEVGPRFCLNPIKIFAGSFGGPTLYENPLYVSPNQIRALEKRNKA 285
Query 121 LDFA 124
FA
Sbjct 286 GKFA 289
> SPBC800.06
Length=295
Score = 99.8 bits (247), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 54/135 (40%), Positives = 76/135 (56%), Gaps = 8/135 (5%)
Query 2 FGVEFDREPHFRVLKELFMQVFGTPRNHPRSKPFFDHAMSFFVTDNRIWFRHYQIAPLVM 61
F FD PH +V+KEL Q FG P+ RSKPF D + + D +IWFR+Y+I
Sbjct 151 FDKTFDTAPHLKVVKELLQQTFGIPKGARRSKPFIDRVCTLTIADGKIWFRNYEIR---- 206
Query 62 GEGGD-ANNPHGQTFIEIGPRFVLDPVKILEGSFCGKTLWRNAEFVRTRNLMRPRRLREA 120
E D + +P T IEIGPRFV+ + ILEGSF G +++N FV + + R + A
Sbjct 207 -ENEDKSKDP--VTLIEIGPRFVMTIINILEGSFGGPVIYKNDTFVSSTMVRAAIRNQAA 263
Query 121 LDFARRQGQKEKRRL 135
+ RQ K +R++
Sbjct 264 QRYVNRQESKLERQV 278
> YOL077c
Length=291
Score = 99.0 bits (245), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 52/149 (34%), Positives = 88/149 (59%), Gaps = 9/149 (6%)
Query 2 FGVEFDREPHFRVLKELFMQVFGTPRNHPRSKPFFDHAMSFFVTDNRIWFRHYQIAPLVM 61
F F+ PH++++KEL + F P N +SKPF DH MSF + D++IW R Y+I+
Sbjct 139 FDQRFESSPHYQLIKELLVHNFCVPPNARKSKPFIDHVMSFSIVDDKIWVRTYEISHSTK 198
Query 62 GEGGDANNPHGQTFIEIGPRFVLDPVKILEGSFCGKTLWRNAEFVRTRNLMRPRRLREAL 121
+ + + +EIGPRFV+ + ILEGSF G ++ N ++V + N++R + ++A
Sbjct 199 NKEEYEDGEEDISLVEIGPRFVMTVILILEGSFGGPKIYENKQYV-SPNVVRAQIKQQAA 257
Query 122 DFARRQGQ-----KEKRRLYLESMLESEP 145
+ A+ + + K KRR E++L ++P
Sbjct 258 EEAKSRAEAAVERKIKRR---ENVLAADP 283
> CE00268
Length=352
Score = 87.4 bits (215), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 43/116 (37%), Positives = 65/116 (56%), Gaps = 13/116 (11%)
Query 2 FGVEFDREPHFRVLKELFMQVFGTPRNHPRSKPFFDHAMSFFVTD-NRIWFRHYQIAPLV 60
F FD++P +++K + MQ GTP +HPRS+PF DH +F V + ++IWFR++QI
Sbjct 171 FDDAFDKKPQLKLIKAVLMQTLGTPHHHPRSQPFVDHVFNFSVGEGDKIWFRNFQIVDES 230
Query 61 MGEGGDANNPHGQTFIEIGPRFVLDPVKILEGSFCGKTLWRNAEFVRTRNLMRPRR 116
+ E+GPRFVL+ V++ GSF G L+ N +V + R R
Sbjct 231 L------------QLQEVGPRFVLEMVRLFAGSFEGAVLYDNPNYVSPNVIRREHR 274
> ECU01g0360
Length=196
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 33/101 (32%), Positives = 47/101 (46%), Gaps = 22/101 (21%)
Query 1 IFGVEFDREPHFRVLKELFMQVFGTPRNHPRSKPFFDHAMSFFVTDNRIWFRHYQIAPLV 60
+F EF+ H + K++ +F RS D A+ FF D IW R Y+I
Sbjct 105 MFSKEFEDIEHLKYAKDVIEHIF-------RSNETKDKALCFFYLDGVIWVRCYKI---- 153
Query 61 MGEGGDANNPHGQTFIEIGPRFVLDPVKILEGSFCGKTLWR 101
G+ EIGPR VL+ +K+LE F GK L++
Sbjct 154 -----------GKKLEEIGPRLVLEVLKVLEKCFEGKALYK 183
> CE19462
Length=149
Score = 34.7 bits (78), Expect = 0.085, Method: Compositional matrix adjust.
Identities = 21/66 (31%), Positives = 40/66 (60%), Gaps = 6/66 (9%)
Query 97 KTLWRNAEFVRTRNLMRPRRLREALDFARRQGQKEKRRLYLESMLESE-PRNKLATEE-- 153
K ++N+EF+R N + R + +++A++Q +K KR++ L S + E NK A+++
Sbjct 7 KFFFKNSEFIR--NFLEIRVIIRNMEYAKKQAEKHKRKIILPSYISWEYGSNKFASQKGQ 64
Query 154 -VFGTD 158
FGT+
Sbjct 65 PAFGTN 70
> SPAC6B12.01
Length=361
Score = 33.1 bits (74), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 13/26 (50%), Positives = 17/26 (65%), Gaps = 0/26 (0%)
Query 75 FIEIGPRFVLDPVKILEGSFCGKTLW 100
IEIGPR L+ +KI E + GK L+
Sbjct 261 LIEIGPRMTLELIKITEDAMGGKVLY 286
> 7300704
Length=460
Score = 31.2 bits (69), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 14/50 (28%), Positives = 23/50 (46%), Gaps = 0/50 (0%)
Query 53 HYQIAPLVMGEGGDANNPHGQTFIEIGPRFVLDPVKILEGSFCGKTLWRN 102
H +A + +G + EIGPR + +KI EG G+ L+ +
Sbjct 248 HVVLAQTLKSKGNLEDKKSSIKLHEIGPRLTMQLIKIEEGLLTGEVLYHD 297
> ECU08g1130
Length=177
Score = 30.4 bits (67), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 25/80 (31%), Positives = 36/80 (45%), Gaps = 14/80 (17%)
Query 15 LKELFMQVFGTPRNHPRSKPFFDHAMSFFVTDNRIWFRHYQIAPLVMGEGGDANNPHGQT 74
LKELF+ + P+ R+ + F+ + I RHY I + E D
Sbjct 111 LKELFLSLSSRPKTFKRN-------LHFYFDGDLIHVRHYAI---LTKEEEDIK----VG 156
Query 75 FIEIGPRFVLDPVKILEGSF 94
F EIGPR + VK ++G F
Sbjct 157 FHEIGPRITMRLVKKMDGFF 176
> Hs6042196
Length=484
Score = 30.0 bits (66), Expect = 1.8, Method: Composition-based stats.
Identities = 20/66 (30%), Positives = 34/66 (51%), Gaps = 12/66 (18%)
Query 100 WRNAEFVRTRNLMRPRRLREALDFARRQ---------GQKEKRRLYLESMLESEPRNKLA 150
WR + FV N++R +++ +ALD Q ++E R +YL ++L EP NK+
Sbjct 206 WRLSVFVN--NMVRAQKI-QALDRGTAQYGVTKFSDLTEEEFRTIYLNTLLRKEPGNKMK 262
Query 151 TEEVFG 156
+ G
Sbjct 263 QAKSVG 268
> At1g79420
Length=404
Score = 29.3 bits (64), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 22/63 (34%), Positives = 31/63 (49%), Gaps = 12/63 (19%)
Query 82 FVLDPVKILEGSFCGKTLWRNAEFVRTRNLMRP----RRLREALD-------FARRQGQK 130
VL K++ GS GKT+WR+ ++ T P RRL + LD FA+ Q
Sbjct 185 LVLGGTKLISGSD-GKTVWRHTPWLGTHAAKGPQRPLRRLIQGLDPKTTASLFAKAQCLG 243
Query 131 EKR 133
E+R
Sbjct 244 ERR 246
> CE28332
Length=384
Score = 29.3 bits (64), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 24/80 (30%), Positives = 37/80 (46%), Gaps = 10/80 (12%)
Query 39 AMSFFVTDNRIWFRHYQIAPLVMGEGGDANNPHGQTFIEIGPRFVLDPVKILEGSFCGKT 98
++F + I+FRH++ EG A +E+GPRF L + +G+F K
Sbjct 312 VVTFHNQRDYIFFRHHRYE--FKKEGSKA------ALLELGPRFTLRLKWLQKGTFDAK- 362
Query 99 LWRNAEFVRTRNLMRPRRLR 118
W E+V R+ M R R
Sbjct 363 -WGEFEWVLKRHEMETSRRR 381
> 7291614
Length=873
Score = 29.3 bits (64), Expect = 3.7, Method: Composition-based stats.
Identities = 37/161 (22%), Positives = 66/161 (40%), Gaps = 20/161 (12%)
Query 9 EPHFRVLKELFMQVFGTPRNHPRSKPFFDHAMSFF----VTDNRIWFRH------YQIAP 58
E F+ +++L+ + P+ K F D SF NR+ Y++
Sbjct 240 EEQFKQVRQLY-----EINDDPKRKEFLDDLFSFMQKRGTPINRLPIMAKSVLDLYELYN 294
Query 59 LVMGEGGDANNPHGQTFIEIGPRFVLDPVKILEGSFCGKTLWRNAEF---VRTRNLMRPR 115
LV+ GG + + + + EI L P I +F +T + + +NL P
Sbjct 295 LVIARGGLVDVINKKLWQEIIKGLHL-PSSITSAAFTLRTQYMKYLYPYECEKKNLSTPA 353
Query 116 RLREALDFARRQGQKEKRRLYLESMLESEPRNKLATEEVFG 156
L+ A+D RR+G++ Y E+M P ++ + G
Sbjct 354 ELQAAIDGNRREGRRSSYGQY-EAMHNQMPMTPISRPSLPG 393
> Hs18572525
Length=338
Score = 28.9 bits (63), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 34/135 (25%), Positives = 56/135 (41%), Gaps = 15/135 (11%)
Query 10 PHFRVLKELFMQVFGTPRNHPRSKPFFDHAMSFF----VTDNRIWFRHYQIAPL------ 59
PH +E F Q++ P+ K F D SF NR+ Q+ L
Sbjct 21 PHEWTYEEQFKQLYEL-DADPKRKEFLDDLFSFMQKRGTPVNRVPIMAKQVLDLYALFRL 79
Query 60 VMGEGGDANNPHGQTFIEIGPRFVLDPVKILEGSFCGKTLWRNAEF---VRTRNLMRPRR 116
V +GG + + + E+ R + P I +F +T + + TR L P
Sbjct 80 VTAKGGLVEVINRKVWREVT-RGLSLPTTITSAAFTLRTQYMKYLYPYECETRALSSPGE 138
Query 117 LREALDFARRQGQKE 131
L+ A+D RR+G+++
Sbjct 139 LQAAIDSNRREGRRQ 153
> CE18041_1
Length=400
Score = 28.5 bits (62), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 22/63 (34%), Positives = 33/63 (52%), Gaps = 10/63 (15%)
Query 77 EIGPRFVLDPVKILEGSFCGKTLW--RNA----EFVRTRNLMRPRRLREALDFARRQGQK 130
EIGPR L+ VKI EG G+ L+ NA E ++ R M ++ + RR+ +
Sbjct 342 EIGPRLTLELVKIEEGIDEGEVLYHKHNAKTPDELIKLRAHMDKKKQMK----KRREQES 397
Query 131 EKR 133
E+R
Sbjct 398 EQR 400
> CE18013_2
Length=140
Score = 28.5 bits (62), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 12/33 (36%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 71 HGQTFIEIGPRFVLDPVKILEGSFCGKTLWRNA 103
HG +I + V DP K ++G +C T ++NA
Sbjct 52 HGSFWIGVYRPTVYDPFKTMDGRYCQYTHFKNA 84
> 7300800
Length=739
Score = 28.1 bits (61), Expect = 6.8, Method: Composition-based stats.
Identities = 17/37 (45%), Positives = 20/37 (54%), Gaps = 1/37 (2%)
Query 116 RLREALDFARRQGQKEKRRLYLESMLESEPRNKLATE 152
RL+ A+D R E R LE LE+E R KLA E
Sbjct 416 RLKRAIDSVRGDNDSEALRAQLEYHLEAE-RRKLAVE 451
> Hs9910566
Length=473
Score = 27.7 bits (60), Expect = 10.0, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 19/43 (44%), Gaps = 0/43 (0%)
Query 60 VMGEGGDANNPHGQTFIEIGPRFVLDPVKILEGSFCGKTLWRN 102
V G G EIGPR L +K+ EG GK ++ +
Sbjct 257 VAGRGNMRAQQSAVRLTEIGPRMTLQLIKVQEGVGEGKVMFHS 299
Lambda K H
0.326 0.142 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2208717646
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40