bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1221_orf1
Length=444
Score E
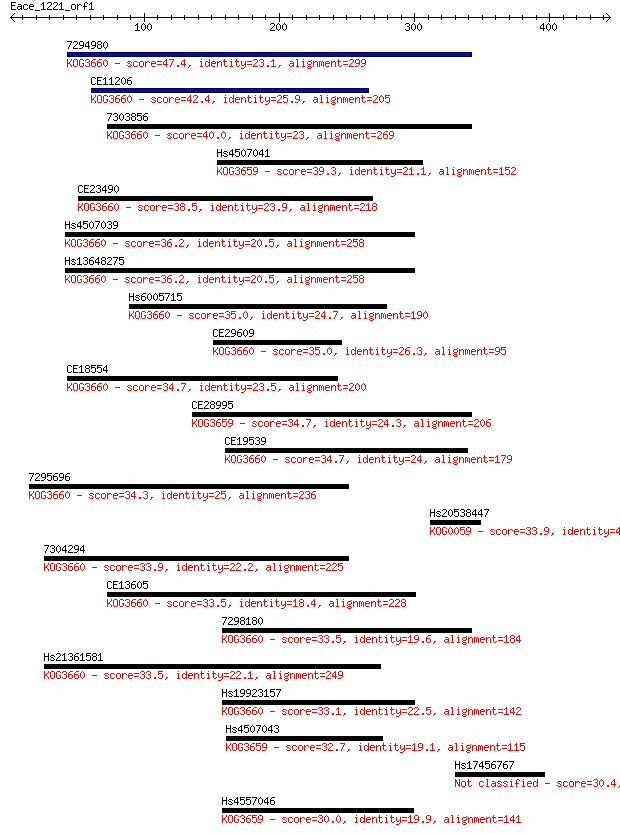
Sequences producing significant alignments: (Bits) Value
7294980 47.4 6e-05
CE11206 42.4 0.002
7303856 40.0 0.011
Hs4507041 39.3 0.018
CE23490 38.5 0.029
Hs4507039 36.2 0.16
Hs13648275 36.2 0.16
Hs6005715 35.0 0.29
CE29609 35.0 0.33
CE18554 34.7 0.39
CE28995 34.7 0.42
CE19539 34.7 0.47
7295696 34.3 0.55
Hs20538447 33.9 0.71
7304294 33.9 0.79
CE13605 33.5 0.96
7298180 33.5 1.0
Hs21361581 33.5 1.1
Hs19923157 33.1 1.1
Hs4507043 32.7 1.6
Hs17456767 30.4 8.3
Hs4557046 30.0 9.7
> 7294980
Length=593
Score = 47.4 bits (111), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 69/304 (22%), Positives = 125/304 (41%), Gaps = 18/304 (5%)
Query 43 ATLHYLEELQPSTLDSSLDPDALRPKALA-TLAFVWLYILAYMWRGLFKAGFPSAILMII 101
+T YL+ + + DS + +LA L WL + + RG+ +G + +L +
Sbjct 163 STQLYLQRIVLNETDSLEEGIGYPSGSLALMLGISWLTVTLIIIRGVKSSGKAAYVLALF 222
Query 102 AMVLCATL--AAVSFFVGTDGSVHVASAISATFDWRPLIHGFFDAAYSASLVRYICQDLC 159
V+ L A++ DG ++ T W L+ Y+A + +C
Sbjct 223 PYVVMFILLVRALTLPGAYDGVMYFL-----TPQWEKLLEP--QVWYNAVTQVFFSLAVC 275
Query 160 LGCCILGTVSSYTKIGFNTYPSGVFTCIVEFLMALVPFVSLLSIVGNLQAVTDIPLAEII 219
G I+ SSY + G N Y ++ +L+ V + I+GNL + +
Sbjct 276 FGVIIM--YSSYNRFGHNVYRDANIVTTLDTFTSLLSGVIIFGILGNLAHESGTKDIASV 333
Query 220 RKTGTAPAYFVVFPAAFEKVSS-PYLFGLFFYGGVFLINTQVAAIGIHSVFNAVLESRLV 278
K G A F+ +P A K P +F L F+ +F++ + +G+ S VL+ + V
Sbjct 334 VKAGPGLA-FISYPDAIAKFKMFPQVFSLLFFAMLFMLGVG-SNVGMVSCIMTVLKDQFV 391
Query 279 N-NCWRRVTSTVIVMICYCISLPFCANNQEVRLQYMSYIVEWLLVPISVFCTTFYVGWVM 337
N W V S + +I + + L + + + M + + +S VGW+
Sbjct 392 NVKLWIIVVS--LSVIGFLVGLIYITPGGQHIITLMDFHGVTFVSLVSAIFELIAVGWIY 449
Query 338 GFQR 341
G +R
Sbjct 450 GTKR 453
> CE11206
Length=768
Score = 42.4 bits (98), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 53/214 (24%), Positives = 96/214 (44%), Gaps = 18/214 (8%)
Query 61 DPDALRPKALATLAFVWLYILAYMWRGLFKAGFPSAILMIIAMVLCATLA--AVSFFVGT 118
D +++ L +A W+ + RG+ G S + +I+ + +L VS
Sbjct 313 DFNSINLPILGAVALCWVIAAVVLIRGMKAIGKLSYVTVILPYFIIVSLLICGVSLPGAK 372
Query 119 DGSVHVASAISATFDWRPLIHGFFDAAYSASLVRYICQDLCLGCCILGTVSSYTKIGFNT 178
DG H+ + ++ ++A+L + IC L +G L ++SY K +N
Sbjct 373 DGLYHLFGQTDFNYLYK-------RETWTAAL-QQICFSLSIGQGGLMNIASYNKKTYNW 424
Query 179 YPSGVFTCIVEFLMALVPFVSLLSIVGNLQ-----AVTDIPLAEIIRKTGTAPAYFVVFP 233
Y ++ LM+++ +++ S +G L TDI I K G A A F+V+
Sbjct 425 YRDVYIIVFLDTLMSILGGITVFSTLGFLAYERGINSTDIDAFNHIVKEGHALA-FIVYT 483
Query 234 AAFEKVSSPYLFGLFFYGGVFL--INTQVAAIGI 265
A ++ PYL+ F+ +FL I+T+V + I
Sbjct 484 EAIAEMPVPYLWYALFFIMLFLLGISTEVVIVEI 517
> 7303856
Length=651
Score = 40.0 bits (92), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 62/279 (22%), Positives = 118/279 (42%), Gaps = 29/279 (10%)
Query 73 LAFVWLYILAYMWRGLFKAGFPSAILMIIAMVLCATL--AAVSFFVGTDGSVHVASAISA 130
LA W+ I M++G+ +G S L + V+ L A++ DG ++
Sbjct 253 LAVAWIVIAGIMFKGVKSSGKASYFLALFPYVVMLVLLVRALTLPGAFDGVLYFL----- 307
Query 131 TFDWRPLIHGFFDAA--YSASLVRYICQDLCLGCCILGTVSSYTKIGFNTYPSGVFTCIV 188
RP H + Y+A + +C G I+ +SY + G N Y +
Sbjct 308 ----RPQWHKLLEPQVWYAAVTQVFFSLAICFGNIIM--YASYNRFGHNIYRDANIVTTL 361
Query 189 EFLMALVPFVSLLSIVGNL---QAVTDIPLAEIIRKTGTAPAYFVVFPAAFEKVSS-PYL 244
+ +L+ V + I+GNL TDI A ++ G F+ +P A K P L
Sbjct 362 DTFTSLLSGVIIFGILGNLAYENNTTDI--ASVV--NGGPGLAFISYPDAIAKFKWLPQL 417
Query 245 FGLFFYGGVFLINTQVAAIGIHSVFNAVLESRLVN-NCWRRVTSTVIVMICYCISLPFCA 303
F + F+ +F++ + +G+ S + V++ + + W V I ++ Y + L +
Sbjct 418 FSVLFFLMLFVLGIG-SNVGMASCMSTVIKDQFGHLKNWTVVVG--IAIVGYFLGLLYIT 474
Query 304 NNQEVRLQYMSYI-VEWLLVPISVFCTTFYVGWVMGFQR 341
+ L + Y V ++ + +++F + W+ G +R
Sbjct 475 PGGQFLLNLVDYFGVTFVALVLAIF-ELVTIAWIYGVKR 512
> Hs4507041
Length=620
Score = 39.3 bits (90), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 32/153 (20%), Positives = 66/153 (43%), Gaps = 5/153 (3%)
Query 154 ICQDLCLGCCILGTVSSYTKIGFNTYPSGVFTCIVEFLMALVPFVSLLSIVGNLQAVTDI 213
+C L +G +L SSY K N Y + T + L + + S +G + +
Sbjct 318 VCFSLGVGFGVLIAFSSYNKFTNNCYRDAIVTTSINSLTSFSSGFVVFSFLGYMAQKHSV 377
Query 214 PLAEIIRKTGTAPAY-FVVFPAAFEKVSSPYLFGLFFYGGVFLINTQVAAIGIHSVFNAV 272
P+ ++ + P F+++P A + + + F+ + + A G+ SV +
Sbjct 378 PIGDVAKD---GPGLIFIIYPEAIATLPLSSAWAVVFFIMLLTLGIDSAMGGMESVITGL 434
Query 273 LESRLVNNCWRRVTSTVIVMICYCISLPFCANN 305
++ + + R + + IV+ + +SL FC N
Sbjct 435 IDEFQLLHRHRELFTLFIVLATFLLSL-FCVTN 466
> CE23490
Length=635
Score = 38.5 bits (88), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 52/233 (22%), Positives = 111/233 (47%), Gaps = 27/233 (11%)
Query 51 LQPSTLDSSLDPDALRPKALATLAFVWLYILAYMWRGLFKAGFPS--AILMIIAMVLCAT 108
++PST D +++ LA +A W+ + + RG+ G S +L+ A+++
Sbjct 188 VKPST--GFTDFNSINVPMLAAVAVCWVIAIISLIRGMKAIGKLSYYTVLLPYAIIIALM 245
Query 109 LAAVSFFVGTDGSVHVASAISATFDWRPLIHGFFDAA-------YSASLVRYICQDLCLG 161
+ ++ +G+ ++ + F +P +G +D A + A+L++ +C L +G
Sbjct 246 IRGLTL----EGA---STGLHFLFWGKPNEYGHYDFAGLYDPDVWIAALMQ-MCFSLSIG 297
Query 162 CCILGTVSSYTKIGFNTYPSGVFTCIVEFLMALVPFVSLLSIVGNLQAVTDIPLA----- 216
L + SY K +N Y + + LM+L+ ++ S +G L +I ++
Sbjct 298 QGGLMNIGSYNKKTYNWYRDAYLVVLCDTLMSLLGGTAVFSTLGFLAEQRNISISNPKGF 357
Query 217 -EIIRKTGTAPAYFVVFPAAFEKVSSPYLFGLFFYGGVFLINTQVAAIGIHSV 268
E++++ G A A F+V+ A ++ P+L+ F+ +FL+ + I +V
Sbjct 358 NEVVQE-GHALA-FIVYTEAIAQMPYPFLWYALFFVMLFLLGISTEIVIIETV 408
> Hs4507039
Length=599
Score = 36.2 bits (82), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 53/272 (19%), Positives = 102/272 (37%), Gaps = 36/272 (13%)
Query 42 KATLHYLEELQPSTLDSSLDPDALRPKALATLAFVWLYILAYMWRGL--------FKAGF 93
A + + E D P +R TLA W+ + +W+G+ F A +
Sbjct 187 SAVVEFWERNMHQMTDGLDKPGQIRWPLAITLAIAWILVYFCIWKGVGWTGKVVYFSATY 246
Query 94 PSAILMII---AMVLCATLAAVSFFVGTDGSVHVASAISATFDWRPLIHGFFDAAYSASL 150
P +L+I+ + L + F++ + S + + DAA
Sbjct 247 PYIMLIILFFRGVTLPGAKEGILFYITPNFRKLSDSEV------------WLDAATQI-- 292
Query 151 VRYICQDLCLGCCILGTVSSYTKIGFNTYPSGVFTCIVEFLMALVPFVSLLSIVGNLQAV 210
+ L LG I + SY N Y + C + ++ + SIVG + V
Sbjct 293 --FFSYGLGLGSLI--ALGSYNSFHNNVYRDSIIVCCINSCTSMFAGFVIFSIVGFMAHV 348
Query 211 TDIPLAEIIRKTGTAPAY-FVVFPAAFEKVSSPYLFGLFFYGGVFLINTQVAAIGIHSVF 269
T +A++ + P F+ +P A ++ L+ + F+ + ++ +
Sbjct 349 TKRSIADV---AASGPGLAFLAYPEAVTQLPISPLWAILFFSMLLMLGIDSQFCTVEGFI 405
Query 270 NAVLES--RLVNNCWRRVTSTVIVMICYCISL 299
A+++ RL+ N R + + +I Y I L
Sbjct 406 TALVDEYPRLLRNR-RELFIAAVCIISYLIGL 436
> Hs13648275
Length=599
Score = 36.2 bits (82), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 53/272 (19%), Positives = 102/272 (37%), Gaps = 36/272 (13%)
Query 42 KATLHYLEELQPSTLDSSLDPDALRPKALATLAFVWLYILAYMWRGL--------FKAGF 93
A + + E D P +R TLA W+ + +W+G+ F A +
Sbjct 187 SAVVEFWERNMHQMTDGLDKPGQIRWPLAITLAIAWILVYFCIWKGVGWTGKVVYFSATY 246
Query 94 PSAILMII---AMVLCATLAAVSFFVGTDGSVHVASAISATFDWRPLIHGFFDAAYSASL 150
P +L+I+ + L + F++ + S + + DAA
Sbjct 247 PYIMLIILFFRGVTLPGAKEGILFYITPNFRKLSDSEV------------WLDAATQI-- 292
Query 151 VRYICQDLCLGCCILGTVSSYTKIGFNTYPSGVFTCIVEFLMALVPFVSLLSIVGNLQAV 210
+ L LG I + SY N Y + C + ++ + SIVG + V
Sbjct 293 --FFSYGLGLGSLI--ALGSYNSFHNNVYRDSIIVCCINSCTSMFAGFVIFSIVGFMAHV 348
Query 211 TDIPLAEIIRKTGTAPAY-FVVFPAAFEKVSSPYLFGLFFYGGVFLINTQVAAIGIHSVF 269
T +A++ + P F+ +P A ++ L+ + F+ + ++ +
Sbjct 349 TKRSIADV---AASGPGLAFLAYPEAVTQLPISPLWAILFFSMLLMLGIDSQFCTVEGFI 405
Query 270 NAVLES--RLVNNCWRRVTSTVIVMICYCISL 299
A+++ RL+ N R + + +I Y I L
Sbjct 406 TALVDEYPRLLRNR-RELFIAAVCIISYLIGL 436
> Hs6005715
Length=642
Score = 35.0 bits (79), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 47/201 (23%), Positives = 93/201 (46%), Gaps = 30/201 (14%)
Query 89 FKAGFPSAILMIIAMVLCATL----AAVSFFVGTDGSVHVASAISATFDWRPLIHGFFDA 144
F A FP +L+I+ +V ATL +S+++G + + + W+ DA
Sbjct 267 FTALFPYVVLLIL-LVRGATLEGASKGISYYIGAQSNF---TKLKEAEVWK-------DA 315
Query 145 AYSASLVRYICQDLCLGCCILGTVSSYTKIGFNTYPSGVFTCIVEFLMALVPFVSLLSIV 204
A I L + L +SSY K N + + C+ L ++ ++ SI+
Sbjct 316 ATQ------IFYSLSVAWGGLVALSSYNKFKNNCFSDAIVVCLTNCLTSVFAGFAIFSIL 369
Query 205 GNLQAVTDIPLAEIIRKTGTAPAYFVVFPAAFEKV-SSPYLFGLFFYGGVFL-INTQVAA 262
G++ ++ +++++ K+G A F+ +P A ++ P+ LFF+ + L +++Q A+
Sbjct 370 GHMAHISGKEVSQVV-KSGFDLA-FIAYPEALAQLPGGPFWSILFFFMLLTLGLDSQFAS 427
Query 263 IG-----IHSVFNAVLESRLV 278
I I +F V++ V
Sbjct 428 IETITTTIQDLFPKVMKKMRV 448
> CE29609
Length=728
Score = 35.0 bits (79), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 25/95 (26%), Positives = 51/95 (53%), Gaps = 2/95 (2%)
Query 151 VRYICQDLCLGCCILGTVSSYTKIGFNTYPSGVFTCIVEFLMALVPFVSLLSIVGNLQAV 210
++ +C L +G L ++SSY+ N + + I + M+LV ++ + +G L
Sbjct 366 LKQLCFSLSVGHGGLISLSSYSPKRNNIFRDALIVIIGDTTMSLVGGGAVFATLGYLAKA 425
Query 211 TDIPLAEIIRKTGTAPAYFVVFPAAFEKVSSPYLF 245
T + +++ K+G + A FVV+P A ++ P+L+
Sbjct 426 TGQDVKDVV-KSGLSLA-FVVYPEAMTRMPVPWLW 458
> CE18554
Length=706
Score = 34.7 bits (78), Expect = 0.39, Method: Compositional matrix adjust.
Identities = 47/202 (23%), Positives = 88/202 (43%), Gaps = 12/202 (5%)
Query 43 ATLHYLEELQPSTLDSSLDPDALRPKALATLAFVWLYILAYMWRGLFKAGFPSAILMIIA 102
AT Y + D L+ ++ K+ + WL ++ +W+G+ G S +++ +
Sbjct 234 ATEQYFFDYVVKPTDGFLEFSSINFKSFFGMNACWLIVVLILWKGVGYMGKASYVIVTLP 293
Query 103 MVLCATLAAVSFFVGT--DGSVHVASAISATFDWRPLIHGFFDAAYSASLVRYICQDLCL 160
+ L FF G DG+ DW + F A + +L + +C L +
Sbjct 294 YFIIILL----FFRGITLDGAEDGIYYYLGNPDWSKV---FMPATWGEAL-KQLCFSLGI 345
Query 161 GCCILGTVSSYTKIGFNTYPSGVFTCIVEFLMALVPFVSLLSIVGNLQAVTDIPLAEIIR 220
G L ++S++K N + + I + +M+LV ++ S +G L +AE++
Sbjct 346 GYGGLIGMASFSKPNNNCFRDALIVIIGDTMMSLVGGAAVFSTLGFLAKKKGCSVAEVVN 405
Query 221 KTGTAPAYFVVFPAAFEKVSSP 242
G + A FV FP A ++ P
Sbjct 406 D-GFSLA-FVAFPEALGRMPLP 425
> CE28995
Length=671
Score = 34.7 bits (78), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 50/214 (23%), Positives = 83/214 (38%), Gaps = 17/214 (7%)
Query 136 PLIHGFFDAAYSASLVRYICQDLCLGCCILGTVSSYTKIGFNTYPSGVFTCIVEFLMALV 195
P D A ++ I L G +L +SSY N Y V T I+ +
Sbjct 340 PDFEKLKDPAVWSAAATQIFFSLGPGFGVLLALSSYNDFNNNCYRDAVTTSIINCATSFF 399
Query 196 PFVSLLSIVGNLQAVTDIPLAEIIRKTGTAPAYFVVFPAAFEKVSSPYLFGLFFYGGVFL 255
+ S +G + +T+ P+ E++ + A F+V+P A + + F+ V L
Sbjct 400 SGCVVFSTLGYMSLLTNKPINEVVGEH-DASLIFIVYPQALATMDYSCFWSFIFF--VML 456
Query 256 INTQVAAIGIHSVFNAVL--------ESRLVNNCWRRVTSTVIVMICYCISLPFCANNQE 307
I +GI S F + ESR ++ R+ VI +I Y +S P + +
Sbjct 457 IT-----LGIDSTFAGIEAFITGFCDESRFLSKN-RKWFVLVICIIYYFLSFPAISYGGQ 510
Query 308 VRLQYMSYIVEWLLVPISVFCTTFYVGWVMGFQR 341
+ ++ L V V C V W G +
Sbjct 511 FVIPFLDEYGVSLSVLFIVTCEMIAVCWFYGVDQ 544
> CE19539
Length=559
Score = 34.7 bits (78), Expect = 0.47, Method: Compositional matrix adjust.
Identities = 43/185 (23%), Positives = 82/185 (44%), Gaps = 10/185 (5%)
Query 160 LGCCILG--TVSSYTKIGFNTYPSGVFTCIVEFLMALVPFVSLLSIVGNLQAVTDIPLAE 217
+ CC G T++SY++ N Y IV+ +++LV + S +G I L +
Sbjct 219 VSCCSGGLFTIASYSRFHNNIYKDIWLVLIVDVIVSLVGCLLTFSAIGFTCYEFAISLDK 278
Query 218 IIRKTGTAPAYFVVFPAAFEKVSSPYLFGLFFYGGVFLINTQVAAIGIHSVFNAVLES-- 275
+ G FV A VS L+ F+ + L+ + ++ +++ +
Sbjct 279 FHIRDGFHLV-FVFLAEALAGVSVAPLYAGLFFIMILLVVHATQMFVVETIVSSICDEYP 337
Query 276 -RLVNNCWRRVTSTVIVMICYCISLPFCANNQEVRLQYMS-YIVEWLLVPISVFCTTFYV 333
RL N RR T + + +S+PFC ++ ++ ++ +++ W LV I+ F +
Sbjct 338 ERLRRN--RRHVLTTVCALFILLSIPFCLSSGLFWMELLTQFVLTWPLVVIA-FLECMAI 394
Query 334 GWVMG 338
WV G
Sbjct 395 NWVYG 399
> 7295696
Length=943
Score = 34.3 bits (77), Expect = 0.55, Method: Compositional matrix adjust.
Identities = 59/249 (23%), Positives = 97/249 (38%), Gaps = 33/249 (13%)
Query 15 VDCNAAGHGVLCSYVPDTNICIASP-TGKATLHYLEELQPSTLDSSLD-PDALRPKALAT 72
+DCN + C I ++P T + E + + L+ P +R + A
Sbjct 456 IDCNNRWNTPDCWVPQRKGINASAPDTSRTPSEEFFENKVLQISGGLEYPGMMRWELFAC 515
Query 73 LAFVWLYILAYMWRGL--------FKAGFPSAILMIIAMVLCATLAAVSFFVGTDGSVHV 124
L WL + W+ + F A FP +L+II MV TL DG+
Sbjct 516 LICAWLMVYFATWKSIKSSAKVRYFTATFP-FVLIIILMVRAVTL---------DGA--- 562
Query 125 ASAISATFDWRPLIHGFFDAAY--SASLVRYICQDLCLGCCILGTVSSYTKIGFNTYPSG 182
A F +RP +A +A+ + + G I + +SY K N
Sbjct 563 --AEGLRFFFRPKWSELKNANVWINAASQNFNSLGITFGSMI--SFASYNKYNNNILRDT 618
Query 183 VFTCIVEFLMALVPFVSLLSIVGNLQAVTDIPLAEIIRKTGTAPAY-FVVFPAAFEKVSS 241
V V + +L+ + S +GNL + + ++I G P FVV+P A K+
Sbjct 619 VAVSAVNMITSLLVGIFAFSTLGNLALEQNTNVRDVI---GDGPGMIFVVYPQAMAKMPY 675
Query 242 PYLFGLFFY 250
L+ + F+
Sbjct 676 AQLWAVMFF 684
> Hs20538447
Length=4273
Score = 33.9 bits (76), Expect = 0.71, Method: Composition-based stats.
Identities = 15/43 (34%), Positives = 23/43 (53%), Gaps = 6/43 (13%)
Query 312 YMSYIVEWL------LVPISVFCTTFYVGWVMGFQRQATIVGL 348
Y+ + V WL L P+S+FC+ +V W G Q T+ G+
Sbjct 189 YLVHAVSWLRVYQQVLSPLSMFCSKVFVQWQQGSLLQKTLTGM 231
> 7304294
Length=522
Score = 33.9 bits (76), Expect = 0.79, Method: Compositional matrix adjust.
Identities = 50/236 (21%), Positives = 89/236 (37%), Gaps = 30/236 (12%)
Query 26 CSYVPDTNICIASPTGKATLHYLEELQPSTLDSSLDPDALRPKALATLAFVWLYILAYMW 85
C+ +T C++ K + + + +T + +R + TL VW+ +W
Sbjct 123 CNNWWNTVNCVSQYERKNLHCWDKIINGTTQKVCSEIGNIRWELAGTLLLVWILCYFCIW 182
Query 86 RGL--------FKAGFPSAILMII---AMVLCATLAAVSFFVGTDGSVHVASAISATFDW 134
+G+ F A FP +L I+ + L L + F++ + S S +
Sbjct 183 KGVKWTGKVVYFTALFPYVLLTILLVRGITLPGALEGIKFYIIPNFSKLTNSEV------ 236
Query 135 RPLIHGFFDAAYSASLVRYICQDLCLGCCILGTVSSYTKIGFNTYPSGVFTCIVEFLMAL 194
+ DA V I LG L + SY K N Y + C V ++
Sbjct 237 ------WIDA------VTQIFFSYGLGLGTLVALGSYNKFTNNVYKDALIVCTVNSSTSM 284
Query 195 VPFVSLLSIVGNLQAVTDIPLAEIIRKTGTAPAYFVVFPAAFEKVSSPYLFGLFFY 250
+ S++G + P+A+ + +G A+ V A + SP LFF+
Sbjct 285 FAGFVIFSVIGFMAHEQQRPVAD-VAASGPGLAFLVYPSAVLQLPGSPMWSCLFFF 339
> CE13605
Length=609
Score = 33.5 bits (75), Expect = 0.96, Method: Compositional matrix adjust.
Identities = 42/239 (17%), Positives = 97/239 (40%), Gaps = 31/239 (12%)
Query 73 LAFVWLYILAYMWRGL--------FKAGFPSA---ILMIIAMVLCATLAAVSFFVGTDGS 121
+A WL + +W+G+ F A FP IL+I + L + F++ + +
Sbjct 225 MAAAWLIVYFALWKGITQARKFVYFCALFPYVLIFILLIRGLTLEGAGTGIYFYLKPNAT 284
Query 122 VHVASAISATFDWRPLIHGFFDAAYSASLVRYICQDLCLGCCILGTVSSYTKIGFNTYPS 181
+ +A+ W+ DA + +G L + S+ K N +
Sbjct 285 RLLDTAV-----WK-------DAGTQ------VFYSYGVGFGALIALGSHNKFNHNCFKD 326
Query 182 GVFTCIVEFLMALVPFVSLLSIVGNLQAVTDIPLAEIIRKTGTAPAYFVVFPAAFEKVSS 241
+ C + ++ ++ SI+G + V ++EI+ K G A F+ +P +
Sbjct 327 AITMCFINGCTSITAGFAVFSILGYMSHVAQKDISEIV-KPGVGLA-FLAYPEVASNLPM 384
Query 242 PYLFGLFFYGGVFLINTQVAAIGIHSVFNAVLESRLVNNCWRRVTSTVIVMICYCISLP 300
+F + F+ + ++ + +F A+ ++ + +++ + + + +CI +P
Sbjct 385 KQVFAVLFFLMITILGLDSQVCMMEGLFTALEDAFPILRKYKKQSLGIFCLFFFCIGIP 443
> 7298180
Length=639
Score = 33.5 bits (75), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 36/187 (19%), Positives = 81/187 (43%), Gaps = 10/187 (5%)
Query 158 LCLGCCILGTVSSYTKIGFNTYPSGVFTCIVEFLMALVPFVSLLSIVGNL-QAVTDIPLA 216
+C G I+ SS+ K G N + ++ + +L+ ++ I+G+L + +
Sbjct 324 VCFGNIIM--YSSFNKFGHNVHRDAAIVTGLDTMTSLLAGFTIFGILGHLAHEIGTDDIG 381
Query 217 EIIRKTGTAPAYFVVFPAAFEKVSS-PYLFGLFFYGGVFLINTQVAAIGIHSVFNAVLES 275
+++ G A F+ +P A K + P +F + F+ +F++ + I + S +
Sbjct 382 SVVK--GGAGLAFISYPDAIAKFKNLPQIFSVLFFLMLFVLGIG-SNIAMTSCSVTAIRD 438
Query 276 RLVN-NCWRRVTSTVIVMICYCISLPFCANNQEVRLQYMSYIVEWLLVPISVFCTTFYVG 334
R N W+ S +I ++ + I L + + L + + ++ + + +G
Sbjct 439 RFPNFGQWQ--CSLLIAVVSFFIGLMYITPGGQYMLTLVDFFGASMIALVLGIAELYTIG 496
Query 335 WVMGFQR 341
W+ G R
Sbjct 497 WIYGTDR 503
> Hs21361581
Length=602
Score = 33.5 bits (75), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 55/260 (21%), Positives = 103/260 (39%), Gaps = 28/260 (10%)
Query 26 CSYVPDTNICIASPTGKATLHYLEELQPSTL---DSSLDPDALRPKALATLAFVWLYILA 82
C TN + + AT +E + L D ALR + L W+
Sbjct 162 CMEFQKTNGSLNGTSENATSPVIEFWERRVLKISDGIQHLGALRWELALCLLLAWVICYF 221
Query 83 YMWRGL--------FKAGFPSAILMIIAMVLCATLAAVSFFVGTDGSVHVASAISATFDW 134
+W+G+ F A FP +++++ ++ TL G + + T W
Sbjct 222 CIWKGVKSTGKVVYFTATFPY-LMLVVLLIRGVTLP------GAAQGIQFYLYPNLTRLW 274
Query 135 RPLIHGFFDAAYSASLVRYICQDLCLGCCILGTVSSYTKIGFNTYPSGVFTCIVEFLMAL 194
P + + DA + +CLGC L + SY K N Y + C + +
Sbjct 275 DPQV--WMDAGTQI----FFSFAICLGC--LTALGSYNKYHNNCYRDCIALCFLNSGTSF 326
Query 195 VPFVSLLSIVGNLQAVTDIPLAEIIRKTGTAPAYFVVFPAAFEKVSSPYLFGLFFYGGVF 254
V ++ SI+G + +P++E + ++G A F+ +P A + L+ F+ V
Sbjct 327 VAGFAIFSILGFMSQEQGVPISE-VAESGPGLA-FIAYPRAVVMLPFSPLWACCFFFMVV 384
Query 255 LINTQVAAIGIHSVFNAVLE 274
L+ + + S+ A+++
Sbjct 385 LLGLDSQFVCVESLVTALVD 404
> Hs19923157
Length=614
Score = 33.1 bits (74), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 32/145 (22%), Positives = 62/145 (42%), Gaps = 7/145 (4%)
Query 158 LCLGCCILGTVSSYTKIGFNTYPSGVFTCIVEFLMALVPFVSLLSIVGNLQAVTDIPLAE 217
+C GC L + SY K N Y + C + + V + SI+G + +P++E
Sbjct 297 ICQGC--LTALGSYNKYHNNCYKDCIALCFLNSATSFVAGFVVFSILGFMSQEQGVPISE 354
Query 218 IIRKTGTAPAYFVVFPAAFEKVSSPYLFGLFFYGGVFLINTQVAAIGIHSVFNAVLE--- 274
+ ++G A F+ FP A + L+ F+ + + + + + A ++
Sbjct 355 -VAESGPGLA-FIAFPKAVTMMPLSQLWSCLFFIMLIFLGLDSQFVCVECLVTASIDMFP 412
Query 275 SRLVNNCWRRVTSTVIVMICYCISL 299
+L + R + I ++CY I L
Sbjct 413 RQLRKSGRRELLILTIAVMCYLIGL 437
> Hs4507043
Length=630
Score = 32.7 bits (73), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 22/115 (19%), Positives = 50/115 (43%), Gaps = 1/115 (0%)
Query 161 GCCILGTVSSYTKIGFNTYPSGVFTCIVEFLMALVPFVSLLSIVGNLQAVTDIPLAEIIR 220
G +L +SY K N Y + T +V + + V + +++G + + + ++E+ +
Sbjct 340 GFGVLLAFASYNKFNNNCYQDALVTSVVNCMTSFVSGFVIFTVLGYMAEMRNEDVSEVAK 399
Query 221 KTGTAPAYFVVFPAAFEKVSSPYLFGLFFYGGVFLINTQVAAIGIHSVFNAVLES 275
G + F+ + A + + F + F+ + + G+ V AVL+
Sbjct 400 DAGPS-LLFITYAEAIANMPASTFFAIIFFLMLITLGLDSTFAGLEGVITAVLDE 453
> Hs17456767
Length=879
Score = 30.4 bits (67), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 17/66 (25%), Positives = 35/66 (53%), Gaps = 2/66 (3%)
Query 330 TFYVGWVMGFQRQATIVGLKACILFGGLCSIVGACYIIAVFCCQDSVFLPTWVMIFVNAL 389
TF + + G +R + + C ++ L SI G C I+ + + S+ P ++ + + AL
Sbjct 581 TFLLSGIPGLERMHIWISIPLCFMY--LVSIPGNCTILFIIKTERSLHEPMYLFLSMLAL 638
Query 390 IAVGVA 395
I +G++
Sbjct 639 IDLGLS 644
> Hs4557046
Length=617
Score = 30.0 bits (66), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 28/146 (19%), Positives = 62/146 (42%), Gaps = 7/146 (4%)
Query 158 LCLGCCILGTVSSYTKIGFNTYPSGVFTCIVEFLMALVPFVSLLSIVGNLQAVTDIPLAE 217
L G +L +SY K N Y + T + + + V ++ SI+G + + + +
Sbjct 319 LGAGFGVLIAFASYNKFDNNCYRDALLTSSINCITSFVSGFAIFSILGYMAHEHKVNIED 378
Query 218 IIRKTGTAPAYFVVFPAAFEKVSSPYLFGLFFYGGVFLINTQVAAIGIHSVFNAVLESRL 277
+ T A F+++P A +S + + F+ + + + G+ +V + +
Sbjct 379 V--ATEGAGLVFILYPEAISTLSGSTFWAVVFFVMLLALGLDSSMGGMEAVITGLADDFQ 436
Query 278 VNNCWRR-----VTSTVIVMICYCIS 298
V R+ VT + ++ +CI+
Sbjct 437 VLKRHRKLFTFGVTFSTFLLALFCIT 462
Lambda K H
0.330 0.142 0.455
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 11571344072
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40