bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
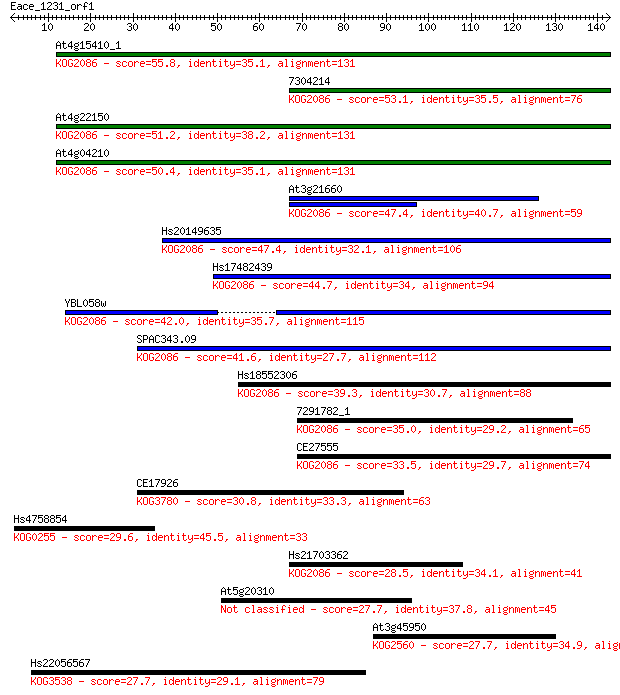
Query= Eace_1231_orf1
Length=142
Score E
Sequences producing significant alignments: (Bits) Value
At4g15410_1 55.8 2e-08
7304214 53.1 2e-07
At4g22150 51.2 6e-07
At4g04210 50.4 1e-06
At3g21660 47.4 8e-06
Hs20149635 47.4 9e-06
Hs17482439 44.7 6e-05
YBL058w 42.0 4e-04
SPAC343.09 41.6 5e-04
Hs18552306 39.3 0.002
7291782_1 35.0 0.051
CE27555 33.5 0.13
CE17926 30.8 0.88
Hs4758854 29.6 1.9
Hs21703362 28.5 3.9
At5g20310 27.7 6.6
At3g45950 27.7 7.3
Hs22056567 27.7 7.4
> At4g15410_1
Length=463
Score = 55.8 bits (133), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 46/178 (25%), Positives = 71/178 (39%), Gaps = 48/178 (26%)
Query 12 IRSLADLRET--QDEPHDDSSGTSSFAGGERSGLAIQNPSAAF----------------- 52
IR+ ADL + E D + GG++SG+ +Q+P
Sbjct 135 IRTFADLNRSPADGEGSDSDEANEYYTGGQKSGMMVQDPKKVKDVDELFDQARQSAVDRP 194
Query 53 --PSATRGAAPAGSRR-------------------------VTLYANGFTVDDGPFREFG 85
PS + + G+ R +T + NGFTV+DGP R F
Sbjct 195 VEPSRSASTSFTGAARLLSGEAVSSSPQQQQQEQPQRIMHTITFWLNGFTVNDGPLRGFS 254
Query 86 RRENDVFIEELKAGVAPKELQQGGRQVHVLLD-DRHEEKYTPPPPPQYVLFGGEGQSL 142
EN F+ + P EL+ +++ V +D R E +T PP P+ F G G++L
Sbjct 255 DPENAAFMNSISRSECPSELEPADKKIPVHVDLVRRGENFTEPPKPKNP-FQGVGRTL 311
> 7304214
Length=407
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 27/76 (35%), Positives = 42/76 (55%), Gaps = 2/76 (2%)
Query 67 VTLYANGFTVDDGPFREFGRRENDVFIEELKAGVAPKELQQGGRQVHVLLDDRHEEKYTP 126
+ L++ GF++D G R + +N F+E + G P+EL + GR V+V ++D E +
Sbjct 217 LKLWSQGFSIDGGELRHYDDPQNKEFLETVMRGEIPQELLEMGRMVNVDVEDHRHEDFKR 276
Query 127 PPPPQYVLFGGEGQSL 142
P PQ F G GQ L
Sbjct 277 QPVPQ--TFKGSGQKL 290
> At4g22150
Length=302
Score = 51.2 bits (121), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 50/177 (28%), Positives = 76/177 (42%), Gaps = 48/177 (27%)
Query 12 IRSLADL-RETQDEPHDDSSGTSS-FAGGERSGLAIQNPSAA------------------ 51
IR+L+DL R ++ + DS G F GGE+SG+ +Q+P+
Sbjct 19 IRTLSDLNRRSEPDSDSDSDGPQEYFTGGEKSGMLVQDPTKEPKHDDVDEIFNQARQLGA 78
Query 52 ------FPSATRGAAPAGS------------------RRVTLYANGFTVDDGPFREFGRR 87
PS++R G + ++NGFTVDDGP R+
Sbjct 79 VEGPLEHPSSSRSFTGTGRLLSGESVPTALQQPEPVIHNIIFWSNGFTVDDGPLRKLDDP 138
Query 88 ENDVFIEELKAGVAPKELQQGGRQ--VHVLLDDRHEEKYTPPPPPQYVLFGGEGQSL 142
EN F++ ++ PKEL+ ++ VHV L R E+ P V F G G++L
Sbjct 139 ENASFLDSIRKSECPKELEPVDKRAPVHVNLMRRDEK--CPEKEKLKVSFQGVGRTL 193
> At4g04210
Length=308
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 46/177 (25%), Positives = 75/177 (42%), Gaps = 46/177 (25%)
Query 12 IRSLADL--RETQDEPHDDSSGTSSFAGGERSGLAIQNPSAAF----------------- 52
IR+L+DL R D D + GGE+SG+ +Q+PS
Sbjct 18 IRTLSDLNRRSGPDSDSDSDGPQEYYTGGEKSGMLVQDPSKKDDVDEIFNQARQLGAVEG 77
Query 53 ----PSATRG-----------AAPAGSRR-------VTLYANGFTVDDGPFREFGRREND 90
P ++R P G+++ + ++NGFT+DDGP R+ EN
Sbjct 78 PLEPPPSSRSFTGTGRLLSGENVPTGNQQPEPVVHNIVFWSNGFTIDDGPLRKLDDPENA 137
Query 91 VFIE-----ELKAGVAPKELQQGGRQVHVLLDDRHEEKYTPPPPPQYVLFGGEGQSL 142
F+E ++ PKEL+ R+ V ++ +E+ P + V F G G++L
Sbjct 138 SFLEVNDFHSIRKSECPKELEPADRRAPVHVNLMRKEEKCPERQKRRVSFQGVGRTL 194
> At3g21660
Length=435
Score = 47.4 bits (111), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 24/59 (40%), Positives = 34/59 (57%), Gaps = 2/59 (3%)
Query 67 VTLYANGFTVDDGPFREFGRRENDVFIEELKAGVAPKELQQGGRQVHVLLDDRHEEKYT 125
VT ++NGFTVDD + EN F+E + + +P+EL Q QV ++ R EE YT
Sbjct 155 VTSWSNGFTVDDSSLKTLDDPENATFLEIISSMESPRELGQVRVQVKII--SREEENYT 211
Score = 38.1 bits (87), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 15/30 (50%), Positives = 20/30 (66%), Gaps = 0/30 (0%)
Query 67 VTLYANGFTVDDGPFREFGRRENDVFIEEL 96
VT++ NGFTVDD PF+ EN F+E +
Sbjct 273 VTIWRNGFTVDDDPFKSLDDPENAAFLEYM 302
> Hs20149635
Length=370
Score = 47.4 bits (111), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 34/109 (31%), Positives = 54/109 (49%), Gaps = 4/109 (3%)
Query 37 GGERSGLAIQNPSAAFPSATRGAAPAGSRRV-TLYANGFTVDDGPFREFGRRENDVFIEE 95
GG R G A + SA R + V L+ +GF++D+G R + N F+E
Sbjct 153 GGYRLGAAPEEESAYVAGEKRQHSSQDVHVVLKLWKSGFSLDNGELRSYQDPSNAQFLES 212
Query 96 LKAGVAPKELQQ--GGRQVHVLLDDRHEEKYTPPPPPQYVLFGGEGQSL 142
++ G P EL++ G QV++ ++D +E + P + F GEGQ L
Sbjct 213 IRRGEVPAELRRLAHGGQVNLDMEDHRDEDFVKPKGA-FKAFTGEGQKL 260
> Hs17482439
Length=175
Score = 44.7 bits (104), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 32/105 (30%), Positives = 50/105 (47%), Gaps = 13/105 (12%)
Query 49 SAAFPSATRGAAPAGSR----------RVTLYANGFTVDDGPFREFGRRENDVFIEELKA 98
S A S + G P G+R + L+ NGFTV++ FR + + F+ +K
Sbjct 38 SRARRSGSEGLQPNGARSQTVLKQVDVNIKLWKNGFTVNN-YFRSYSNGASQQFLNSIKK 96
Query 99 GVAPKELQQ-GGRQVHVLLDDRHEEKYTPPPPPQYVLFGGEGQSL 142
G P ELQ ++V V ++D+ E Y P + LF G+G +
Sbjct 97 GELPSELQGIFDKEVGVKVEDKKNETYMSTKPV-FQLFSGQGHQV 140
> YBL058w
Length=423
Score = 42.0 bits (97), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 27/81 (33%), Positives = 41/81 (50%), Gaps = 3/81 (3%)
Query 64 SRRVTLYANGFTVDDGPFREFGRRENDVFIEELKAGVAPKEL--QQGGRQVHVLLDDRHE 121
+R +T + GF V DGP + N ++ EL G AP +L Q G++V V + + +
Sbjct 234 TREITFWKEGFQVADGPLYRYDDPANSFYLSELNQGRAPLKLLDVQFGQEVEVNVYKKLD 293
Query 122 EKYTPPPPPQYVLFGGEGQSL 142
E Y P + F G+GQ L
Sbjct 294 ESYK-APTRKLGGFSGQGQRL 313
Score = 29.3 bits (64), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 14/36 (38%), Positives = 22/36 (61%), Gaps = 0/36 (0%)
Query 14 SLADLRETQDEPHDDSSGTSSFAGGERSGLAIQNPS 49
S +D+ Q + D+ ++FAGGE SGL + +PS
Sbjct 128 SFSDMVRGQADDDDEDQPRNTFAGGETSGLEVTDPS 163
> SPAC343.09
Length=410
Score = 41.6 bits (96), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 31/114 (27%), Positives = 47/114 (41%), Gaps = 6/114 (5%)
Query 31 GTSSFAGGERSGLAIQNPSAAFPSATRGAAPAGSRRVTLYANGFTVDDGPFREFGRREND 90
GT + A G + + P P R + + NGF+VDDGP + N
Sbjct 180 GTENEASGSTTPVTQSGPPRENPPTESQPEKPLRRTLYFWRNGFSVDDGPIYTYDDPANQ 239
Query 91 VFIEELKAGVAPKEL--QQGGRQVHVLLDDRHEEKYTPPPPPQYVLFGGEGQSL 142
+ + +G AP L + + V++ R +E Y P P F G+GQ L
Sbjct 240 EMLRYINSGRAPLHLLGVSMNQPIDVVVQHRMDEDYVAPFKP----FSGKGQRL 289
> Hs18552306
Length=259
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 27/95 (28%), Positives = 46/95 (48%), Gaps = 9/95 (9%)
Query 55 ATRGAAPAGSRR-----VTLYANGFTVDDGPFREFGRRENDVFIEELKAGVAPKELQQ-- 107
+++ +PA ++ + L+ NGFTV+D FR + + F+ +K G P ELQ
Sbjct 48 SSKCVSPAEQKKQVDVNIKLWKNGFTVNDD-FRSYSDGASQQFLNSIKKGELPSELQGIF 106
Query 108 GGRQVHVLLDDRHEEKYTPPPPPQYVLFGGEGQSL 142
+V V ++D+ E P + F G+G L
Sbjct 107 DKEEVDVKVEDKKNE-ICLSTKPVFQPFSGQGHRL 140
> 7291782_1
Length=788
Score = 35.0 bits (79), Expect = 0.051, Method: Compositional matrix adjust.
Identities = 19/65 (29%), Positives = 34/65 (52%), Gaps = 1/65 (1%)
Query 69 LYANGFTVDDGPFREFGRRENDVFIEELKAGVAPKELQQGGRQVHVLLDDRHEEKYTPPP 128
L++ GF++DDG R + EN+ F+ + G P+E+ + R V + + D E Y
Sbjct 163 LWSEGFSLDDGSLRLYALPENERFLRAILRGDFPEEMLRVPR-VQLSVQDHTNESYRHLS 221
Query 129 PPQYV 133
Q++
Sbjct 222 RKQFM 226
> CE27555
Length=301
Score = 33.5 bits (75), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 22/76 (28%), Positives = 35/76 (46%), Gaps = 6/76 (7%)
Query 69 LYANGFTVDDGPFREFGRRENDVFIEELKAGVAPKELQQG--GRQVHVLLDDRHEEKYTP 126
L+++G +++DGP F+E + G P L Q G+++ ++ HEE P
Sbjct 96 LWSDGLSIEDGPLMSRQDPRTIEFLESVGKGEIPPSLVQQYPGKEIDFKVNRHHEEYVAP 155
Query 127 PPPPQYVLFGGEGQSL 142
P FGG G L
Sbjct 156 KMKP----FGGSGVRL 167
> CE17926
Length=397
Score = 30.8 bits (68), Expect = 0.88, Method: Compositional matrix adjust.
Identities = 21/65 (32%), Positives = 31/65 (47%), Gaps = 3/65 (4%)
Query 31 GTSSFAGGERSGLAIQNPSAAFPSATRGAAPAGSRRVTLYAN--GFTVDDGPFREFGRRE 88
GT F G S + + S +P +T G AP + + + FTVD P+ FG +
Sbjct 282 GTPVFLGNYGSPNSTRR-SIMYPESTSGEAPPSTTYYCVPPSEMSFTVDPPPYSTFGAKT 340
Query 89 NDVFI 93
+ VFI
Sbjct 341 DPVFI 345
> Hs4758854
Length=594
Score = 29.6 bits (65), Expect = 1.9, Method: Composition-based stats.
Identities = 15/34 (44%), Positives = 22/34 (64%), Gaps = 1/34 (2%)
Query 2 LFICCVLLMAIRSLAD-LRETQDEPHDDSSGTSS 34
+F+CCVL + SL+ L ET+D+P +S SS
Sbjct 542 IFLCCVLAIVAFSLSSLLPETRDQPLSESLNHSS 575
> Hs21703362
Length=472
Score = 28.5 bits (62), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 14/41 (34%), Positives = 21/41 (51%), Gaps = 0/41 (0%)
Query 67 VTLYANGFTVDDGPFREFGRRENDVFIEELKAGVAPKELQQ 107
+ LY NG + DGPF+ F + ++ G P ELQ+
Sbjct 202 LKLYRNGIMMFDGPFQPFYDPSTQRCLRDILDGFFPSELQR 242
> At5g20310
Length=394
Score = 27.7 bits (60), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 17/45 (37%), Positives = 23/45 (51%), Gaps = 8/45 (17%)
Query 51 AFPSATRGAAPAGSRRVTLYANGFTVDDGPFREFGRRENDVFIEE 95
PS++ G AP R +NG D+ P RE+DVF+EE
Sbjct 171 VLPSSSSGTAPLSPR-----SNG---DEAPSEMSLSREDDVFLEE 207
> At3g45950
Length=385
Score = 27.7 bits (60), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 15/43 (34%), Positives = 24/43 (55%), Gaps = 5/43 (11%)
Query 87 RENDVFIEELKAGVAPKELQQGGRQVHVLLDDRHEEKYTPPPP 129
R+ E KAG+AP E+ +GG+++++ H KY PP
Sbjct 15 RKQKELEEARKAGLAPAEVDEGGKEINL-----HIPKYLTIPP 52
> Hs22056567
Length=703
Score = 27.7 bits (60), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 23/85 (27%), Positives = 38/85 (44%), Gaps = 13/85 (15%)
Query 6 CVLLMAIRSLADLRETQDEPHDDSSGTSSFAGGERSGLAIQNPS----AAFPSA--TRGA 59
C++++ + SL Q E D+ GT S L + PS A++ A R A
Sbjct 19 CLVVLGLGSLVFATPQQQEYVVDTDGTCSC-------LPVSPPSLPGNASWKKAGVDRAA 71
Query 60 APAGSRRVTLYANGFTVDDGPFREF 84
PA S ++ + G +DD P ++
Sbjct 72 PPASSHPISSHGWGLCLDDPPAKDI 96
Lambda K H
0.317 0.137 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1603110344
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40