bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
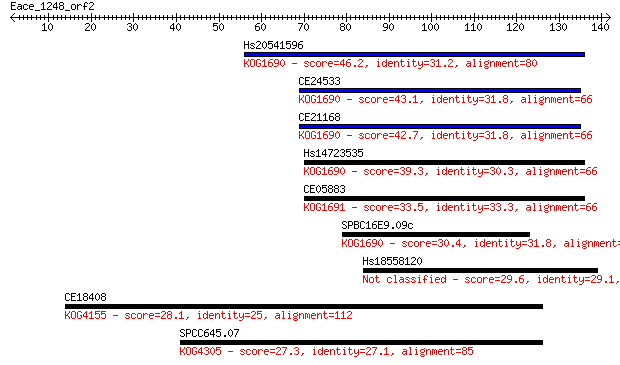
Query= Eace_1248_orf2
Length=141
Score E
Sequences producing significant alignments: (Bits) Value
Hs20541596 46.2 2e-05
CE24533 43.1 2e-04
CE21168 42.7 2e-04
Hs14723535 39.3 0.003
CE05883 33.5 0.13
SPBC16E9.09c 30.4 1.3
Hs18558120 29.6 2.1
CE18408 28.1 6.1
SPCC645.07 27.3 9.4
> Hs20541596
Length=227
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 25/91 (27%), Positives = 40/91 (43%), Gaps = 11/91 (12%)
Query 56 GICGSPLLRAAAAFIFALACVARSASGAYFYVQESQDKCFIESVPTGVALTASYKNH--- 112
G+ PL + LA A A G YF++ E++ +CFIE +P + +Y+
Sbjct 3 GVGAGPLRAMGRQALLLLALCATGAQGLYFHIGETEKRCFIEEIPDETMVIGNYRTQMWD 62
Query 113 --------ENPGVTCSIIFKDPSGRSAYSRE 135
PG+ + KDP G+ SR+
Sbjct 63 KQKEVFLPSTPGLGMHVEVKDPDGKVVLSRQ 93
> CE24533
Length=211
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 21/76 (27%), Positives = 35/76 (46%), Gaps = 10/76 (13%)
Query 69 FIFALACVARSASGAYFYVQESQDKCFIESVPTGVALTASYKNH----------ENPGVT 118
+ AL +A A YF++ E++ KCFIE +P +T +YK + P +
Sbjct 3 LLIALLSLATYADSLYFHIAETEKKCFIEEIPDETMVTGNYKVQLYDPNTKGYGDYPNIG 62
Query 119 CSIIFKDPSGRSAYSR 134
+ KDP + S+
Sbjct 63 MHVEVKDPEDKVILSK 78
> CE21168
Length=167
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 21/76 (27%), Positives = 35/76 (46%), Gaps = 10/76 (13%)
Query 69 FIFALACVARSASGAYFYVQESQDKCFIESVPTGVALTASYKNH----------ENPGVT 118
+ AL +A A YF++ E++ KCFIE +P +T +YK + P +
Sbjct 3 LLIALLSLATYADSLYFHIAETEKKCFIEEIPDETMVTGNYKVQLYDPNTKGYGDYPNIG 62
Query 119 CSIIFKDPSGRSAYSR 134
+ KDP + S+
Sbjct 63 MHVEVKDPEDKVILSK 78
> Hs14723535
Length=214
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 20/77 (25%), Positives = 35/77 (45%), Gaps = 11/77 (14%)
Query 70 IFALACVARSASGAYFYVQESQDKCFIESVPTGVALTASY------KNHEN-----PGVT 118
+ + +A S YF++ E++ KCFIE +P + +Y K E PG+
Sbjct 4 LLLVLWLATRGSALYFHIGETEKKCFIEEIPDETMVIGNYRTQLYDKQREEYQPATPGLG 63
Query 119 CSIIFKDPSGRSAYSRE 135
+ KDP + +R+
Sbjct 64 MFVEVKDPEDKVILARQ 80
> CE05883
Length=204
Score = 33.5 bits (75), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 28/66 (42%), Gaps = 2/66 (3%)
Query 70 IFALACVARSASGAYFYVQESQDKCFIESVPTGVALTASYKNHENPGVTCSIIFKDPSGR 129
IF A V A FYV + KC E + V +T Y+ + T SI D G
Sbjct 6 IFLSAIVL--AHSLRFYVPPKEKKCLKEEIHKNVVVTGEYEFSQGIQYTGSIHVTDTRGH 63
Query 130 SAYSRE 135
+ Y RE
Sbjct 64 TLYKRE 69
> SPBC16E9.09c
Length=215
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 14/49 (28%), Positives = 24/49 (48%), Gaps = 5/49 (10%)
Query 79 SASGAYFYVQESQDKCFIESVPTGVALT-----ASYKNHENPGVTCSII 122
+ +FY S+ KCF++ +P G+ +T + N N VT + I
Sbjct 16 QSQAVHFYFDGSKPKCFLKDMPKGMMVTGIVKAEQWNNEANKWVTSNNI 64
> Hs18558120
Length=101
Score = 29.6 bits (65), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 16/55 (29%), Positives = 25/55 (45%), Gaps = 0/55 (0%)
Query 84 YFYVQESQDKCFIESVPTGVALTASYKNHENPGVTCSIIFKDPSGRSAYSREVLL 138
YF+ E ++KC IE +P+ +T + GV I PS R + +L
Sbjct 17 YFHAGEREEKCLIEDIPSDTLVTEPWGEGIVVGVQVQIGDCFPSKRLVKAANTML 71
> CE18408
Length=1101
Score = 28.1 bits (61), Expect = 6.1, Method: Composition-based stats.
Identities = 28/113 (24%), Positives = 41/113 (36%), Gaps = 13/113 (11%)
Query 14 ALRPPLCCRSPGSAAYILGHSVACLNTCYSVSFPAHSMRRYMGICGSPLLRAAAAFIFAL 73
++PP+ P + ++GH +FP G P+ RA + +
Sbjct 799 GVKPPIFV--PPTGRIMVGHCSLSNKEMKIWAFPEE---------GPPVTRAKLSHSDEI 847
Query 74 ACVARSASGAYFYVQESQDKCF-IESVPTGVALTASYKNHENPGVTCSIIFKD 125
C A S G F S+D I + G LT HEN C I F +
Sbjct 848 TCFATSPKGGNFIATGSRDMSLKIWQIDKGF-LTQVLVGHENVVTCCCISFDE 899
> SPCC645.07
Length=1334
Score = 27.3 bits (59), Expect = 9.4, Method: Composition-based stats.
Identities = 23/85 (27%), Positives = 38/85 (44%), Gaps = 15/85 (17%)
Query 41 CYSVSFPAHSMRRYMGICGSPLLRAAAAFIFALACVARSASGAYFYVQESQDKCFIESVP 100
CYS+S P RR L + F A +S+S A+ + + K +I SVP
Sbjct 564 CYSISCP----RR--------LEQQHRLFAKMRANTEQSSSLAF---DDKEQKLWIHSVP 608
Query 101 TGVALTASYKNHENPGVTCSIIFKD 125
+A + S + + V C +I+ +
Sbjct 609 QEIAYSVSDRERKRQEVICEVIYTE 633
Lambda K H
0.324 0.135 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1608078824
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40