bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1249_orf3
Length=290
Score E
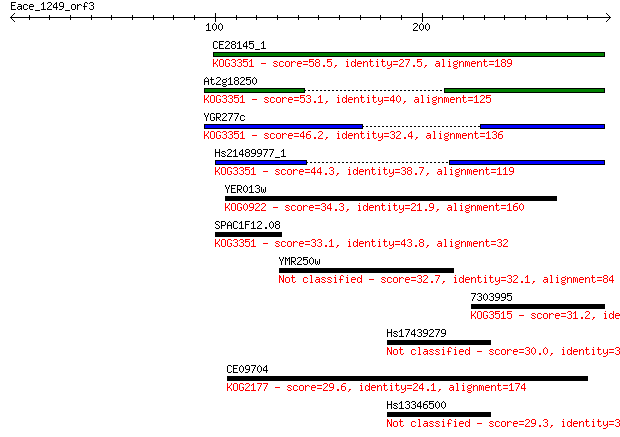
Sequences producing significant alignments: (Bits) Value
CE28145_1 58.5 1e-08
At2g18250 53.1 6e-07
YGR277c 46.2 8e-05
Hs21489977_1 44.3 3e-04
YER013w 34.3 0.35
SPAC1F12.08 33.1 0.75
YMR250w 32.7 0.85
7303995 31.2 2.4
Hs17439279 30.0 6.3
CE09704 29.6 8.5
Hs13346500 29.3 9.3
> CE28145_1
Length=246
Score = 58.5 bits (140), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 52/190 (27%), Positives = 71/190 (37%), Gaps = 67/190 (35%)
Query 99 LVAGTFDRLHAGHQLLLAAAALSAQQRIGVAVASGPLIKRKMASQENVAAAGIEHFAFRL 158
++ GTFDRLH GH++LL AA A I V V +I +K + IE FR+
Sbjct 103 VLGGTFDRLHNGHKVLLNKAAELASDVIVVGVTDKDMIIKKSLFE------MIEPVEFRI 156
Query 159 QAAVAFVQLVAASRGRAVRLTGFKEALVEEEASLSDELQLQSSHLGGSNQFGRDFSSVVE 218
+ V FV+ D S +
Sbjct 157 KKVVDFVE---------------------------------------------DISGTAK 171
Query 219 AFTSPVEEALGDSLQLEVFRITDAVGPADRIA-FDCLVVSDETVKGADMVNRIRGEAGNE 277
P+ D GP+ RI + +VVS ET+KGAD VN+ R E G
Sbjct 172 CLAEPI---------------IDPFGPSTRIKDLEAIVVSRETIKGADAVNKKRNEQGMS 216
Query 278 PVSVLTVGLV 287
+ V+ V LV
Sbjct 217 QLDVIIVELV 226
> At2g18250
Length=176
Score = 53.1 bits (126), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 24/48 (50%), Positives = 32/48 (66%), Gaps = 0/48 (0%)
Query 95 FGYTLVAGTFDRLHAGHQLLLAAAALSAQQRIGVAVASGPLIKRKMAS 142
FG ++ GTFDRLH GH++ L AAA A+ RI V V GP++ +K S
Sbjct 15 FGAVVLGGTFDRLHDGHRMFLKAAAELARDRIVVGVCDGPMLTKKQFS 62
Score = 29.6 bits (65), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 26/89 (29%), Positives = 42/89 (47%), Gaps = 17/89 (19%)
Query 211 RDFSSVVEAFTSPVEE----------ALGDSLQLEVFRITDAVGPA--DRIAFDCLVVSD 258
+ FS +++ P+EE ++ L ++ ITD GP+ D + +VVS
Sbjct 59 KQFSDMIQ----PIEERMRNVETYVKSIKPELVVQAEPITDPYGPSIVDE-NLEAIVVSK 113
Query 259 ETVKGADMVNRIRGEAGNEPVSVLTVGLV 287
ET+ G VNR R E G + + V +V
Sbjct 114 ETLPGGLSVNRKRAERGLSQLKIEVVEIV 142
> YGR277c
Length=305
Score = 46.2 bits (108), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 24/61 (39%), Positives = 36/61 (59%), Gaps = 1/61 (1%)
Query 228 LGDSLQLEVFRITDAVGPADRIA-FDCLVVSDETVKGADMVNRIRGEAGNEPVSVLTVGL 286
L L +E+ + D GP ++ +CLVVS ETV GA+ VN+ R E G P++V V +
Sbjct 209 LKPDLSVELVPLRDVCGPTGKVPEIECLVVSRETVSGAETVNKTRIEKGMSPLAVHVVNV 268
Query 287 V 287
+
Sbjct 269 L 269
Score = 40.4 bits (93), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 20/76 (26%), Positives = 36/76 (47%), Gaps = 6/76 (7%)
Query 95 FGYTLVAGTFDRLHAGHQLLLAAAALSAQQRIGVAVASGPLIKRKMASQENVAAAGIEHF 154
F T + GTFD +H GH++LL+ + QR+ + L++ K + IE +
Sbjct 142 FHVTALGGTFDHIHDGHKILLSVSTFITSQRLICGITCDELLQNKKYKE------LIEPY 195
Query 155 AFRLQAAVAFVQLVAA 170
R + F++L+
Sbjct 196 DTRCRHVHQFIKLLKP 211
> Hs21489977_1
Length=339
Score = 44.3 bits (103), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 20/44 (45%), Positives = 30/44 (68%), Gaps = 0/44 (0%)
Query 100 VAGTFDRLHAGHQLLLAAAALSAQQRIGVAVASGPLIKRKMASQ 143
V GTFDRLH H++LL+ A + AQ+++ V VA L+K K+ +
Sbjct 196 VGGTFDRLHNAHKVLLSVACILAQEQLVVGVADKDLLKSKLLPE 239
Score = 35.4 bits (80), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 26/82 (31%), Positives = 42/82 (51%), Gaps = 7/82 (8%)
Query 213 FSSVVEAFTSPVE---EALGD---SLQLEVFRITDAVGPA-DRIAFDCLVVSDETVKGAD 265
+++ +T VE E L D SL +V + D GPA + + LVVS+ET +G
Sbjct 237 LPELLQPYTERVEHLSEFLVDIKPSLTFDVIPLLDPYGPAGSDPSLEFLVVSEETYRGGM 296
Query 266 MVNRIRGEAGNEPVSVLTVGLV 287
+NR R E E +++ + L+
Sbjct 297 AINRFRLENDLEELALYQIQLL 318
> YER013w
Length=1145
Score = 34.3 bits (77), Expect = 0.35, Method: Composition-based stats.
Identities = 35/165 (21%), Positives = 66/165 (40%), Gaps = 13/165 (7%)
Query 105 DRLHAGHQLLLAAAALSAQQRIGVAVA-----SGPLIKRKMASQENVAAAGIEHFAFRLQ 159
D + G + + + +I +++ SG + KR S E+ + H + ++
Sbjct 224 DVVRQGQHIFVEVIKIQNNGKISLSMKNIDQHSGEIRKRNTESVEDRGRSNDAHTSRNMK 283
Query 160 AAVAFVQLVAASRGRAVRLTGFKEALVEEEASLSDELQLQSSHLGGSNQFGRDFSSVVEA 219
+ L + R +L A +++ L DE+ + +S+L RD S+V
Sbjct 284 NKIKRRALTSPERWEIRQLIASGAASIDDYPELKDEIPINTSYLTAK----RDDGSIVNG 339
Query 220 FTSPVEEALGDSLQLEVFRITDAVGPADRIAFDCLVVSDETVKGA 264
T V+ L + + E I + D F + D+ VKGA
Sbjct 340 NTEKVDSKLEEQQRDETDEIDVELNTDDGPKF----LKDQQVKGA 380
> SPAC1F12.08
Length=329
Score = 33.1 bits (74), Expect = 0.75, Method: Compositional matrix adjust.
Identities = 14/32 (43%), Positives = 19/32 (59%), Gaps = 0/32 (0%)
Query 100 VAGTFDRLHAGHQLLLAAAALSAQQRIGVAVA 131
V GTFD LH GH++LL A + + V V+
Sbjct 161 VGGTFDHLHVGHKVLLTLTAWFGVKEVIVGVS 192
> YMR250w
Length=585
Score = 32.7 bits (73), Expect = 0.85, Method: Compositional matrix adjust.
Identities = 27/87 (31%), Positives = 42/87 (48%), Gaps = 8/87 (9%)
Query 131 ASGPLIKRKMASQENVAAAGIEHFAFRLQAAVAFVQLVAASRGRAVRLT--GFKEALVEE 188
ASG I +E++ A G+E + F V+ + G LT G L +
Sbjct 279 ASGGFIIPFGFEKEHMKAYGMERWGFNHPRVVSM-----NTSGHKFGLTTPGLGWVLWRD 333
Query 189 EASLSDELQLQSSHLGGSNQ-FGRDFS 214
E+ L+DEL+ + +LGG + FG +FS
Sbjct 334 ESLLADELRFKLKYLGGVEETFGLNFS 360
> 7303995
Length=729
Score = 31.2 bits (69), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 33/64 (51%), Gaps = 4/64 (6%)
Query 224 VEEALGDSLQLEVFRITDAVGPADRIAFDCLVVSDETVKGADMVNRIRGEAGNEPVSVLT 283
VEE DSL E I D V PAD A++C+ ++ + V +R E ++P L+
Sbjct 507 VEERKIDSLTYESTLIVDKVAPADYGAYECVARNEL----GEAVETVRLEITSQPDPPLS 562
Query 284 VGLV 287
+ ++
Sbjct 563 LNIL 566
> Hs17439279
Length=212
Score = 30.0 bits (66), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 15/50 (30%), Positives = 24/50 (48%), Gaps = 0/50 (0%)
Query 183 EALVEEEASLSDELQLQSSHLGGSNQFGRDFSSVVEAFTSPVEEALGDSL 232
+ L+E L +ELQL+ S Q D ++E+ S + E +SL
Sbjct 85 QELIETNQQLRNELQLEQSRAANQEQRANDLEQIMESVKSKIGELEDESL 134
> CE09704
Length=944
Score = 29.6 bits (65), Expect = 8.5, Method: Composition-based stats.
Identities = 42/187 (22%), Positives = 75/187 (40%), Gaps = 32/187 (17%)
Query 106 RLHAGHQLLLAAAALSAQQRIGVAVASGPLIKRKMASQENVAAAGIEHFAFRLQA----- 160
RLH GHQL+ A S R +K+K++ + A +E+ RLQ+
Sbjct 202 RLHNGHQLVEKKALTSDDVR---------EMKQKISDASHAAFQSLENLKPRLQSVGGTI 252
Query 161 -AVAFVQLVAASRGRAVRLTGFKEALVEEEASLSDELQLQSSHLGGSNQFGRDFSSVVEA 219
A A +L + + L+ F + +E S DEL + ++ + V+
Sbjct 253 EAKALDKLTSLIKIFEFMLSTFDSKI--KENSTMDELMEEVKKAEKISKVYSEAGGRVDV 310
Query 220 FTSPVEEALG------DSLQLEV-FRITDAVGPADRIAFDCLVVSDETVKGADMVNRIRG 272
+ +E A + L+ E+ F+I + G ++I + GA+ VN +
Sbjct 311 MLTAIESAFNSYWDPFEKLKQEMGFQIDEPTGSVNQIP--------ASEGGAERVNEVAA 362
Query 273 EAGNEPV 279
A P+
Sbjct 363 PAAGRPI 369
> Hs13346500
Length=597
Score = 29.3 bits (64), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 15/50 (30%), Positives = 24/50 (48%), Gaps = 0/50 (0%)
Query 183 EALVEEEASLSDELQLQSSHLGGSNQFGRDFSSVVEAFTSPVEEALGDSL 232
+ L+E L +ELQL+ S Q D ++E+ S + E +SL
Sbjct 85 QELIETNQQLRNELQLEQSRAANQEQRANDLEQIMESVKSKIGELEDESL 134
Lambda K H
0.318 0.131 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6481502418
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40