bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1266_orf3
Length=855
Score E
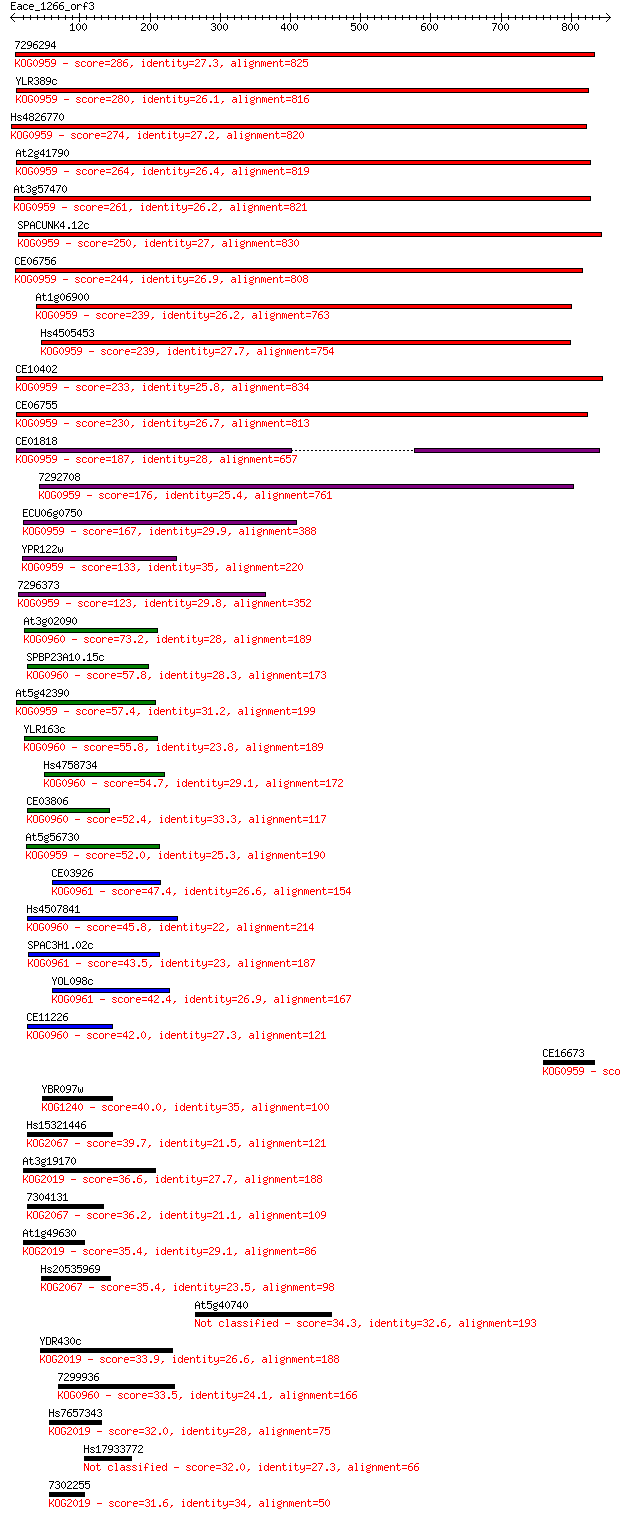
Sequences producing significant alignments: (Bits) Value
7296294 286 1e-76
YLR389c 280 9e-75
Hs4826770 274 5e-73
At2g41790 264 6e-70
At3g57470 261 5e-69
SPACUNK4.12c 250 1e-65
CE06756 244 5e-64
At1g06900 239 1e-62
Hs4505453 239 2e-62
CE10402 233 1e-60
CE06755 230 1e-59
CE01818 187 1e-46
7292708 176 2e-43
ECU06g0750 167 1e-40
YPR122w 133 2e-30
7296373 123 1e-27
At3g02090 73.2 3e-12
SPBP23A10.15c 57.8 1e-07
At5g42390 57.4 2e-07
YLR163c 55.8 4e-07
Hs4758734 54.7 9e-07
CE03806 52.4 5e-06
At5g56730 52.0 5e-06
CE03926 47.4 1e-04
Hs4507841 45.8 4e-04
SPAC3H1.02c 43.5 0.002
YOL098c 42.4 0.005
CE11226 42.0 0.006
CE16673 40.0 0.022
YBR097w 40.0 0.025
Hs15321446 39.7 0.032
At3g19170 36.6 0.23
7304131 36.2 0.37
At1g49630 35.4 0.53
Hs20535969 35.4 0.54
At5g40740 34.3 1.3
YDR430c 33.9 1.4
7299936 33.5 2.0
Hs7657343 32.0 5.8
Hs17933772 32.0 5.9
7302255 31.6 7.5
> 7296294
Length=990
Score = 286 bits (733), Expect = 1e-76, Method: Compositional matrix adjust.
Identities = 225/846 (26%), Positives = 391/846 (46%), Gaps = 53/846 (6%)
Query 8 SDILKPEADYRDFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLE 67
++I K D RD+R QL NG+ + + P ++ + A++ G + DP ++PGLAHF E
Sbjct 25 NNIEKSLQDTRDYRGLQLENGLKVLLISDPNTDVSAAALSVQVGHMSDPTNLPGLAHFCE 84
Query 68 HMLFLGTSKYPEPESYDSFLTQNGGANNAYTDEEKTVFFNKVTDSAFEEALDRFAEFFKS 127
HMLFLGT KYP Y ++L+Q+GG++NA T T + V + ALDRFA+FF +
Sbjct 85 HMLFLGTEKYPHENGYTTYLSQSGGSSNAATYPLMTKYHFHVAPDKLDGALDRFAQFFIA 144
Query 128 PLFNRQYEEKEVNAIDAEHQKNIPNDDERAWYTIRSLAK--GPMSRFATGNAETLSTYPK 185
PLF E+E+NA+++EH+KN+P+D R R LAK S+F +GN TLS PK
Sbjct 145 PLFTPSATEREINAVNSEHEKNLPSDLWRIKQVNRHLAKPDHAYSKFGSGNKTTLSEIPK 204
Query 186 ANGIDLVDRLRDFHSKYYCGSNMVAVTISPRSLDEQESLIREKLEGISAGHADWLGMVQC 245
+ ID+ D L FH ++Y + M I SLDE E ++ EK I + V+
Sbjct 205 SKNIDVRDELLKFHKQWYSANIMCLAVIGKESLDELEGMVLEKFSEIENKN------VKV 258
Query 246 PG-PMFDTVKPFDHTNSGKFIHLQSFSSQPSLWVAFGLPATLTSYKKQPTSVLTYLLEYT 304
PG P P+ G+ + + SL ++F YK P + LT+L+ +
Sbjct 259 PGWPR----HPYAEERYGQKVKIVPIKDIRSLTISFTTDDLTQFYKSGPDNYLTHLIGHE 314
Query 305 GEGSLAKRLRLLGLADGISPAVDRNSIS--TLLGIKVDLTQKGAAHRGLVLQEIFSYINF 362
G+GS+ LR LG + + A +N+ + I VDLTQ+G H +++ +F Y+
Sbjct 315 GKGSILSELRRLGWCNDLM-AGHQNTQNGFGFFDIVVDLTQEGLEHVDDIVKIVFQYLEM 373
Query 363 LRDHGVGHDLVSTMAQQSHIDFH---TTQPSSSIMDEAARLAHNLLTYEPYHVVAGDSLL 419
LR G + + + + F QP + + + + + V+ L
Sbjct 374 LRKEGPKKWIFDECVKLNEMRFRFKEKEQPENLVTHAVSSMQ----IFPLEEVLIAPYLS 429
Query 420 IDADPRLTNQLLQAMSPSKAIIAFSDPDFTSKVDNFETDPYYGVQFRVLDLPQHHAVAMA 479
+ P L LL + PSK+ I F D E PYY ++ + + + +
Sbjct 430 NEWRPDLIKGLLDELVPSKSRIVIVSQSFEPDCDLAE--PYYKTKYGITRVAKDTVQSWE 487
Query 480 VLTASPNAFRMPPPLMHIPKASELKILPGLLGLTEPELISEQGGNAGTAVWWQGQGAFAL 539
+ N ++ P IP ++ +P P +I + VW + F
Sbjct 488 NCELNEN-LKLALPNSFIPTNFDISDVPA-DAPKHPTIILD---TPILRVWHKQDNQFNK 542
Query 540 PRIAVQLNGSISKEKADVLSRTQGSLALAAIAEHLQEETVDFQNCGITHSLAFKGTGFHM 599
P+ + + S D L+ + + + + L E D + + S+ K G
Sbjct 543 PKACMTFDMSNPIAYLDPLNCNLNHMMVMLLKDQLNEYLYDAELASLKLSVMGKSCGIDF 602
Query 600 TFEGYTQAQ---LGKVMAHVASLLSDPSMVEPERFERIKEKQIKLLADPATSMAFEHALE 656
T G++ Q L K++ H+ ++ +RF+ +KE+ ++ L + ++H++
Sbjct 603 TIRGFSDKQVVLLEKLLDHLFDF-----SIDEKRFDILKEEYVRSLKNFKAEQPYQHSIY 657
Query 657 AAAILTRNDAFSRKDVLNALQQSNYEDSIAKLSE-LKNIHVDAFVMGNIDRDQSLAM--- 712
A+L +A++ ++L+A++ Y+ + E + +H + F+ GN+ + Q+ +
Sbjct 658 YLALLLTENAWANMELLDAMELVTYDRVLNFAKEFFQRLHTECFIFGNVTKQQATDIAGR 717
Query 713 VEDFLEQAGFTPIAHDDAVASLAMEQKQTIEATLANPIKGDKDH----ASLVQFQLGIPS 768
V LE + + + + M +K+ + + +K++ +S Q L +
Sbjct 718 VNTRLEATNASKL----PILARQMLKKREYKLLAGDSYLFEKENEFHKSSCAQLYLQCGA 773
Query 769 IEDRVNLAV--LTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTHPDE 826
D N+ V ++Q L+ YD LRT+ QLGYI + + ++ V+ AK HP
Sbjct 774 QTDHTNIMVNLVSQVLSEPCYDCLRTKEQLGYIVFSGVRKVNGANGIRIIVQSAK-HPSY 832
Query 827 VVKMID 832
V I+
Sbjct 833 VEDRIE 838
> YLR389c
Length=988
Score = 280 bits (716), Expect = 9e-75, Method: Compositional matrix adjust.
Identities = 213/835 (25%), Positives = 398/835 (47%), Gaps = 50/835 (5%)
Query 9 DILKPEADYRDFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEH 68
D LKP+ D R +R +L N + A+ + P++++A ++ N G+ DP+++PGLAHF EH
Sbjct 63 DFLKPDLDERSYRFIELPNKLKALLIQDPKADKAAASLDVNIGAFEDPKNLPGLAHFCEH 122
Query 69 MLFLGTSKYPEPESYDSFLTQNGGANNAYTDEEKTVFFNKVTDSAFEEALDRFAEFFKSP 128
+LF+G+ K+P+ Y S+L+++GG++NAYT + T +F +V ALDRF+ FF P
Sbjct 123 LLFMGSEKFPDENEYSSYLSKHGGSSNAYTASQNTNYFFEVNHQHLFGALDRFSGFFSCP 182
Query 129 LFNRQYEEKEVNAIDAEHQKNIPNDDERAWYTIRSL--AKGPMSRFATGNAETLSTYPKA 186
LFN+ +KE+NA+++E++KN+ ND R + +SL K P +F+TGN ETL T PK
Sbjct 183 LFNKDSTDKEINAVNSENKKNLQNDIWRIYQLDKSLTNTKHPYHKFSTGNIETLGTLPKE 242
Query 187 NGIDLVDRLRDFHSKYYCGSNMVAVTISPRSLDEQESLIREKLEGISAGHADWLGMVQCP 246
NG+++ D L FH +Y +N++ + I L RE L+ +S D V
Sbjct 243 NGLNVRDELLKFHKNFY-SANLMKLCI----------LGREDLDTLSDWTYDLFKDVANN 291
Query 247 G---PMF-DTVKPFDHTNSGKFIHLQSFSSQPSLWVAFGLPATLTSYKKQPTSVLTYLLE 302
G P++ + + +H K I ++ L ++F +P ++ +P +L++L+
Sbjct 292 GREVPLYAEPIMQPEHLQ--KIIQVRPVKDLKKLEISFTVPDMEEHWESKPPRILSHLIG 349
Query 303 YTGEGSLAKRLRLLGLADGISPAVDRNSI-STLLGIKVDLTQKGAAHRGLVLQEIFSYIN 361
+ G GSL L+ LG A+ +S S + + +DLT G H V+ IF YI
Sbjct 350 HEGSGSLLAHLKKLGWANELSAGGHTVSKGNAFFAVDIDLTDNGLTHYRDVIVLIFQYIE 409
Query 362 FLRDHGVGHDLVSTMAQQSHIDFHTTQ---PSSSIMDEAARLAHNLLTYEPYHVVAGDSL 418
L++ + + + S+ F Q PSS++ A L + Y P + L
Sbjct 410 MLKNSLPQKWIFNELQDISNATFKFKQAGSPSSTVSSLAKCLEKD---YIPVSRILAMGL 466
Query 419 LIDADPRLTNQLLQAMSPSKAIIAFSDPDFTSKVDNFETDPYYGVQFRVLDLPQHHAVAM 478
L +P L Q A+ P + + + ++ + +YG ++V+D P M
Sbjct 467 LTKYEPDLLTQYTDALVPENSRVTL----ISRSLETDSAEKWYGTAYKVVDYPADLIKNM 522
Query 479 AVLTASPNAFRMPPPLMHIPKASELKILPGLLGLTEPELISEQGGNAGTAVWWQGQGAFA 538
+P A +P P + ++ + G+ L EP L+ + + +W++ F
Sbjct 523 KSPGLNP-ALTLPRPNEFVSTNFKVDKIDGIKPLDEPVLLL---SDDVSKLWYKKDDRFW 578
Query 539 LPRIAVQLNGSISKEKADVLSRTQGSLALAAIAEHLQEETVDFQNCGITHSLAFKGTGFH 598
PR + L+ + A +++ +L + L++ D + S G
Sbjct 579 QPRGYIYLSFKLPHTHASIINSMLSTLYTQLANDALKDVQYDAACADLRISFNKTNQGLA 638
Query 599 MTFEGYTQAQLGKVMAHVASLLSDPSMVEP--ERFERIKEKQIKLLADPATSMAFEHALE 656
+T G+ + K++ + L + EP +RFE +K+K I+ L + + +
Sbjct 639 ITASGFNE----KLIILLTRFLQGVNSFEPKKDRFEILKDKTIRHLKNLLYEVPYSQMSN 694
Query 657 AAAILTRNDAFSRKDVLNALQQSNYEDSIAKLSEL-KNIHVDAFVMGNIDRDQSL---AM 712
+ ++S + L ++ +E I + + + ++ + + GNI +++L ++
Sbjct 695 YYNAIINERSWSTAEKLQVFEKLTFEQLINFIPTIYEGVYFETLIHGNIKHEEALEVDSL 754
Query 713 VEDFLEQAGFTPIAHDDAVASLAMEQKQTIEATLANPIKGDKDHASLVQF--QLGIPSIE 770
++ + ++ + S + + +T A +K ++ S +Q QL + S E
Sbjct 755 IKSLIPNNIHNLQVSNNRLRSYLLPKGKTFRYETA--LKDSQNVNSCIQHVTQLDVYS-E 811
Query 771 DRVNLA-VLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTHP 824
D L+ + Q ++ +D+LRT+ QLGY+ + TA ++ ++ T P
Sbjct 812 DLSALSGLFAQLIHEPCFDTLRTKEQLGYVVFSSSLNNHGTANIRILIQSEHTTP 866
> Hs4826770
Length=1019
Score = 274 bits (701), Expect = 5e-73, Method: Compositional matrix adjust.
Identities = 223/838 (26%), Positives = 381/838 (45%), Gaps = 47/838 (5%)
Query 2 AIGALASDILKPEADYRDFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPG 61
AI + + I K D R++R +L+NG+ + + P ++++ A+ + GSL DP ++ G
Sbjct 46 AIKRIGNHITKSPEDKREYRGLELANGIKVLLMSDPTTDKSSAALDVHIGSLSDPPNIAG 105
Query 62 LAHFLEHMLFLGTSKYPEPESYDSFLTQNGGANNAYTDEEKTVFFNKVTDSAFEEALDRF 121
L+HF EHMLFLGT KYP+ Y FL+++ G++NA+T E T ++ V+ E ALDRF
Sbjct 106 LSHFCEHMLFLGTKKYPKENEYSQFLSEHAGSSNAFTSGEHTNYYFDVSHEHLEGALDRF 165
Query 122 AEFFKSPLFNRQYEEKEVNAIDAEHQKNIPNDDERAWYTIRSLA--KGPMSRFATGNAET 179
A+FF PLF+ +++EVNA+D+EH+KN+ ND R + ++ K P S+F TGN T
Sbjct 166 AQFFLCPLFDESCKDREVNAVDSEHEKNVMNDAWRLFQLEKATGNPKHPFSKFGTGNKYT 225
Query 180 LSTYPKANGIDLVDRLRDFHSKYYCGSNMVAVTISPR-SLDEQESLIREKLEGISAGHAD 238
L T P GID+ L FHS YY SN++AV + R SLD+ +L+ + + +
Sbjct 226 LETRPNQEGIDVRQELLKFHSAYY-SSNLMAVCVLGRESLDDLTNLVVKLFSEVENKNV- 283
Query 239 WLGMVQCPGPMFDTVKPFDHTNSGKFIHLQSFSSQPSLWVAFGLPATLTSYKKQPTSVLT 298
P P F PF + + + +L+V F +P YK P L
Sbjct 284 -------PLPEFPE-HPFQEEHLKQLYKIVPIKDIRNLYVTFPIPDLQKYYKSNPGHYLG 335
Query 299 YLLEYTGEGSLAKRLRLLGLADGISPAVDRNSISTLLG-IKVDLTQKGAAHRGLVLQEIF 357
+L+ + G GSL L+ G + + + + I VDLT++G H ++ +F
Sbjct 336 HLIGHEGPGSLLSELKSKGWVNTLVGGQKEGARGFMFFIINVDLTEEGLLHVEDIILHMF 395
Query 358 SYINFLRDHGVGHDLVSTMAQQSHIDFHTTQPSSSIMDEAARLAHNLLTYEPYHVVAGDS 417
YI LR G + + + F +++A L Y V+ +
Sbjct 396 QYIQKLRAEGPQEWVFQECKDLNAVAFRFKDKERP-RGYTSKIAGILHYYPLEEVLTAEY 454
Query 418 LLIDADPRLTNQLLQAMSPSKAIIAFSDPDFTSKVDNFETDPYYGVQFRVLDLPQHHAVA 477
LL + P L +L + P +A F K D T+ +YG Q++ A+
Sbjct 455 LLEEFRPDLIEMVLDKLRPENVRVAIVSKSFEGKTDR--TEEWYGTQYK------QEAIP 506
Query 478 MAVLTASPNA-----FRMPPPLMHIPKASELKILPGLLGLTE-PELISEQGGNAGTAVWW 531
V+ NA F++P IP + +ILP T P LI + + +W+
Sbjct 507 DEVIKKWQNADLNGKFKLPTKNEFIP--TNFEILPLEKEATPYPALIKD---TVMSKLWF 561
Query 532 QGQGAFALPRIAVQLNGSISKEKADVLSRTQGSLALAAIAEHLQEETVDFQNCGITHSLA 591
+ P+ + D L L L + + L E + G+++ L
Sbjct 562 KQDDKKKKPKACLNFEFFSPFAYVDPLHCNMAYLYLELLKDSLNEYAYAAELAGLSYDLQ 621
Query 592 FKGTGFHMTFEGYTQAQ---LGKVMAHVASLLSDPSMVEPERFERIKEKQIKLLADPATS 648
G +++ +GY Q L K++ +A+ ++ +RFE IKE ++ L +
Sbjct 622 NTIYGMYLSVKGYNDKQPILLKKIIEKMATF-----EIDEKRFEIIKEAYMRSLNNFRAE 676
Query 649 MAFEHALEAAAILTRNDAFSRKDVLNALQQSNYEDSIAKLSE-LKNIHVDAFVMGNIDRD 707
+HA+ +L A+++ ++ AL A + + L +H++A + GNI +
Sbjct 677 QPHQHAMYYLRLLMTEVAWTKDELKEALDDVTLPRLKAFIPQLLSRLHIEALLHGNITKQ 736
Query 708 QSLA---MVED-FLEQAGFTPIAHDDAVASLAMEQKQTIEATLANPIKGDKDHASLVQFQ 763
+L MVED +E A P+ V ++ + + + +Q
Sbjct 737 AALGIMQMVEDTLIEHAHTKPLLPSQLVRYREVQLPDRGWFVYQQRNEVHNNCGIEIYYQ 796
Query 764 LGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAK 821
+ S + + L + Q ++ +++LRT+ QLGYI + +A L+ ++ K
Sbjct 797 TDMQSTSENMFLELFCQIISEPCFNTLRTKEQLGYIVFSGPRRANGIQSLRFIIQSEK 854
> At2g41790
Length=970
Score = 264 bits (675), Expect = 6e-70, Method: Compositional matrix adjust.
Identities = 216/851 (25%), Positives = 382/851 (44%), Gaps = 72/851 (8%)
Query 9 DILKPEADYRDFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEH 68
+ILKP D R++R L N + + + P +++ +++ + GS DP+ + GLAHFLEH
Sbjct 14 EILKPRTDNREYRMIVLKNLLQVLLISDPDTDKCAASMSVSVGSFSDPQGLEGLAHFLEH 73
Query 69 MLFLGTSKYPEPESYDSFLTQNGGANNAYTDEEKTVFFNKVTDSAFEEALDRFAEFFKSP 128
MLF + KYPE +SY ++T++GG+ NAYT E+T + V F+EALDRFA+FF P
Sbjct 74 MLFYASEKYPEEDSYSKYITEHGGSTNAYTASEETNYHFDVNADCFDEALDRFAQFFIKP 133
Query 129 LFNRQYEEKEVNAIDAEHQKNIPNDDERAWYTIRSLAK--GPMSRFATGNAETLSTYPKA 186
L + +E+ A+D+E+QKN+ +D R + L+K P +F+TGN +TL P+A
Sbjct 134 LMSADATMREIKAVDSENQKNLLSDGWRIRQLQKHLSKEDHPYHKFSTGNMDTLHVRPQA 193
Query 187 NGIDLVDRLRDFHSKYYCGSNMVAVTISPRSLDEQESLIREKLEGISAGHADWLGMVQCP 246
G+D L F+ ++Y + M V SLD+ + L+ + I + + + P
Sbjct 194 KGVDTRSELIKFYEEHYSANIMHLVVYGKESLDKIQDLVERMFQEIQNTNK---VVPRFP 250
Query 247 GPMFDTVKPFDHTNSGKFIHLQSFSSQPSLWVAFGLPATLTSYKKQPTSVLTYLLEYTGE 306
G +P + + L V++ + ++ Y + P+ L +L+ + GE
Sbjct 251 G------QPCTADHLQILVKAIPIKQGHKLGVSWPVTPSIHHYDEAPSQYLGHLIGHEGE 304
Query 307 GSLAKRLRLLGLADGISPAVDRNSIS-TLLGIKVDLTQKGAAHRGLVLQEIFSYINFLRD 365
GSL L+ LG A G+S ++ + + +DLT G H +L +F+YI L+
Sbjct 305 GSLFHALKTLGWATGLSAGEGEWTLDYSFFKVSIDLTDAGHEHMQEILGLLFNYIQLLQQ 364
Query 366 HGVGHDLVSTMAQQSHIDFH---TTQPSSSIMDEAARLAHNLLTYEPYHVVAGDSLLIDA 422
GV + ++ FH P S I+D +A N+ Y + G SL
Sbjct 365 TGVCQWIFDELSAICETKFHYQDKIPPMSYIVD----IASNMQIYPTKDWLVGSSLPTKF 420
Query 423 DPRLTNQLLQAMSPSKAIIAFSDPDFTSKVDNFETDPYYGVQFRVLDLPQHHAVAMAVLT 482
+P + +++ +SPS I + F + D E P+Y + L+ + V +
Sbjct 421 NPAIVQKVVDELSPSNFRIFWESQKFEGQTDKAE--PWYNTAYS-LEKITSSTIQEWVQS 477
Query 483 ASPNAFRMPPPLMHIPKASELK------ILPGLLGLTEPELISEQGGNAGTAVWWQGQGA 536
A +P P + IP LK +P LL T + +W++
Sbjct 478 APDVHLHLPAPNVFIPTDLSLKDADDKETVPVLLRKT-----------PFSRLWYKPDTM 526
Query 537 FALPRIAVQLNG----SISKEKADVLSRTQGSLALAAIAEHLQEETVDFQNCGITHSLAF 592
F+ P+ V+++ ++S A VL+ L + ++L E Q G+ + ++
Sbjct 527 FSKPKAYVKMDFNCPLAVSSPDAAVLTDIFTRL----LMDYLNEYAYYAQVAGLYYGVSL 582
Query 593 KGTGFHMTFEGYT---QAQLGKVMAHVASLLSDPSMVEPERFERIKEKQIKLLADPATSM 649
GF +T GY + L V+ +A+ V+P+RF IKE K +
Sbjct 583 SDNGFELTLLGYNHKLRILLETVVGKIANF-----EVKPDRFAVIKETVTKEYQNYKFRQ 637
Query 650 AFEHALEAAAILTRNDAFSRKDVLNALQQSNYEDSIAKLSE--LKNIHVDAFVMGNIDRD 707
+ A+ +++ ++ + + L+ L ED +AK L ++ ++ GN++ +
Sbjct 638 PYHQAMYYCSLILQDQTWPWTEELDVLSHLEAED-VAKFVPMLLSRTFIECYIAGNVENN 696
Query 708 QSLAMVEDFLEQAGFT---PIAH--------DDAVASLAMEQKQTIEATLANPIKGDKDH 756
++ +MV+ +E F PI + V L K +NP D++
Sbjct 697 EAESMVKH-IEDVLFNDPKPICRPLFPSQHLTNRVVKLGEGMKYFYHQDGSNP--SDENS 753
Query 757 ASLVQFQLGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCF 816
A + Q+ + L + + + LRT QLGYI + + +Q
Sbjct 754 ALVHYIQVHRDDFSMNIKLQLFGLVAKQATFHQLRTVEQLGYITALAQRNDSGIYGVQFI 813
Query 817 VEGAKTHPDEV 827
++ + P +
Sbjct 814 IQSSVKGPGHI 824
> At3g57470
Length=989
Score = 261 bits (667), Expect = 5e-69, Method: Compositional matrix adjust.
Identities = 215/871 (24%), Positives = 385/871 (44%), Gaps = 85/871 (9%)
Query 7 ASDILKPEADYRDFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFL 66
+ILKP D R++R L N + + + P +++ ++ + GS DPE + GLAHFL
Sbjct 14 CGEILKPRTDKREYRRIVLKNSLEVLLISDPETDKCAASMNVSVGSFTDPEGLEGLAHFL 73
Query 67 EHMLFLGTSKYPEPESYDSFLTQNGGANNAYTDEEKTVFFNKVTDSAFEEALDRFAEFFK 126
EHMLF + KYPE +SY ++T++GG+ NAYT E T + + +F EALDRFA+FF
Sbjct 74 EHMLFYASEKYPEEDSYSKYITEHGGSTNAYTSSEDTNYHFDINTDSFYEALDRFAQFFI 133
Query 127 SPLFNRQYEEKEVNAIDAEHQKNIPNDDERAWYTIRSLAK--GPMSRFATGNAETLSTYP 184
PL + +E+ A+D+EHQ N+ +D R + L++ P +F+TGN +TL P
Sbjct 134 QPLMSTDATMREIKAVDSEHQNNLLSDSWRMAQLQKHLSREDHPYHKFSTGNMDTLHVRP 193
Query 185 KANGIDLVDRLRDFHSKYYCGSNMVAVTISPRSLDEQESLIREKLEGISAGHADWLGMVQ 244
+ NG+D L F+ ++Y + M V +LD+ + L+ +GI + G+ +
Sbjct 194 EENGVDTRSELIKFYDEHYSANIMHLVVYGKENLDKTQGLVEALFQGIRNTNQ---GIPR 250
Query 245 CPGPMFDTVKPFDHTNSGKFIHLQSFSSQPSLWVAFGLPATLTSYKKQPTSVLTYLLEYT 304
PG +P + + L V++ + +++ Y++ P L L+ +
Sbjct 251 FPG------QPCTLDHLQVLVKAVPIMQGHELSVSWPVTPSISHYEEAPCRYLGDLIGHE 304
Query 305 GEGSLAKRLRLLGLADGI-SPAVDRNSISTLLGIKVDLTQKGAAHRGLVLQEIFSYINFL 363
GEGSL L++LG A G+ + D + + + +DLT G H +L +F YI L
Sbjct 305 GEGSLFHALKILGWATGLYAGEADWSMEYSFFNVSIDLTDAGHEHMQDILGLLFEYIKVL 364
Query 364 RDHGVGHDLVSTMAQQSHIDFHTTQPSSSIMDEAARLAHNLLTYEPYHVVAGDSLLIDAD 423
+ GV + ++ +FH Q + A ++ N+ Y H + G SL +
Sbjct 365 QQSGVSQWIFDELSAICEAEFH-YQAKIDPISYAVDISSNMKIYPTKHWLVGSSLPSKFN 423
Query 424 PRLTNQLLQAMSPSKA------------IIAFSDPDFTSKVDNFETDPYYGVQFRVLDLP 471
P + ++L +SP+ ++ +PD + +P+Y + + +
Sbjct 424 PAIVQKVLDELSPNNPSVPNVLCSYNLHVVQALNPDCLRPRQTDKVEPWYNTAYSLEKIT 483
Query 472 QHHAVAMAVLTASPNAFRMPPPLMHIPKASELK------ILPGLLGLTEPELISEQGGNA 525
+ + + +A +P P + IP LK I P LL T +
Sbjct 484 K-FTIQEWMQSAPDVNLLLPTPNVFIPTDFSLKDLKDKDIFPVLLRKT-----------S 531
Query 526 GTAVWWQGQGAFALPRIAVQLNG----SISKEKADVLSRTQGSLALAAIAEHLQEE---T 578
+ +W++ F P+ V+++ ++S A VLS + + + ++L E
Sbjct 532 YSRLWYKPDTKFFKPKAYVKMDFNCPLAVSSPDAAVLS----DIFVWLLVDYLNEYALIN 587
Query 579 VDF------QNCGITHSLAFKGTGFHMTFEGYT---QAQLGKVMAHVASLLSDPSMVEPE 629
+D+ Q G+ + L+ GF ++ G+ + L V+ +A V+P+
Sbjct 588 LDYVSAYYAQAAGLDYGLSLSDNGFELSLAGFNHKLRILLEAVIQKIAKF-----EVKPD 642
Query 630 RFERIKEKQIKLLADPATSMAFEHALEAAAILTRNDAFSRKDVLNALQQSNYEDSIAKLS 689
RF IKE K + E A +++ ++ + + L+AL ED +A
Sbjct 643 RFSVIKETVTKAYQNNKFQQPHEQATNYCSLVLQDQIWPWTEELDALSHLEAED-LANFV 701
Query 690 E--LKNIHVDAFVMGNIDRDQSLAMVEDFLEQAGFT---PIAH--------DDAVASLAM 736
L V+ ++ GN+++D++ +MV+ +E FT PI + V L
Sbjct 702 PMLLSRTFVECYIAGNVEKDEAESMVKH-IEDVLFTDSKPICRPLFPSQFLTNRVTELGT 760
Query 737 EQKQTIEATLANPIKGDKDHASLVQFQLGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQL 796
K +N D++ A + Q+ L + + + LRT QL
Sbjct 761 GMKHFYYQEGSN--SSDENSALVHYIQVHKDEFSMNSKLQLFELIAKQDTFHQLRTIEQL 818
Query 797 GYIAGAKESQAASTALLQCFVEGAKTHPDEV 827
GYI S + +Q ++ + P +
Sbjct 819 GYITSLSLSNDSGVYGVQFIIQSSVKGPGHI 849
> SPACUNK4.12c
Length=969
Score = 250 bits (638), Expect = 1e-65, Method: Compositional matrix adjust.
Identities = 224/863 (25%), Positives = 381/863 (44%), Gaps = 62/863 (7%)
Query 13 PEADYRDFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFL 72
P D R++R +L N + + V P ++ A A+ + GS +P ++ GLAHF EH+LF+
Sbjct 17 PNLDDREYRLIKLENDLEVLLVRDPETDNASAAIDVHIGSQSNPRELLGLAHFCEHLLFM 76
Query 73 GTSKYPEPESYDSFLTQNGGANNAYTDEEKTVFFNKVTDSAFEEALDRFAEFFKSPLFNR 132
GT KYP+ Y +L + G +NAYT T ++ +V+ A ALDRFA+FF PLF
Sbjct 77 GTKKYPDENEYRKYLESHNGISNAYTASNNTNYYFEVSHDALYGALDRFAQFFIDPLFLE 136
Query 133 QYEEKEVNAIDAEHQKNIPNDDERAW--YTIRSLAKGPMSRFATGNAETLSTYPKANGID 190
+ +++E+ A+D+EH KN+ +D R W Y++ S K S+F TGN ETL PK G+D
Sbjct 137 ECKDREIRAVDSEHCKNLQSDSWRFWRLYSVLSNPKSVFSKFNTGNIETLGDVPKELGLD 196
Query 191 LVDRLRDFHSKYYCGSNMVAVTISPRSLDEQESLIREKLEGISAGHADWLGMVQCPGPMF 250
+ L F+ KYY + M V I LD + E I P P F
Sbjct 197 VRQELLKFYDKYYSANIMKLVIIGREPLDVLQDWAAELFSPIKNK--------AVPIPKF 248
Query 251 DTVKPFDHTNSGKFIHLQSFSSQPSLWVAFGLPATLTSYKKQPTSVLTYLLEYTGEGSLA 310
P+ K +++ + L + F +P YK +P + +LL + GEGS
Sbjct 249 PD-PPYTDNEVRKICYVKPVKNLRRLDIVFPIPGQYHKYKCRPAEYVCHLLGHEGEGSYL 307
Query 311 KRLRLLGLADG-ISPAVDRNSISTLLGIKVDLTQKGAAHRGLVLQEIFSYINFLRDHGVG 369
L+ LGLA I+ V + ++ + LT++G V++ +F YI L
Sbjct 308 AYLKSLGLATSLIAFNVSITEDADIIVVSTFLTEEGLTDYQRVIKILFEYIRLLDQTNAH 367
Query 370 HDLVSTMAQQSHIDFHTTQPSSSIMDEAARLAHNLLT-----YEPYHVVAGDSLLIDADP 424
L S F T Q + A + AH + + Y V+ S+L + DP
Sbjct 368 KFLFEETRIMSEAQFKTRQKTP-----AYQYAHVVASKLQREYPRDKVLYYSSVLTEFDP 422
Query 425 RLTNQLLQAMSPSKAIIAFSDPDFTSKVDNFETDPYYGVQFRVLDLPQHHAVAMAVLTAS 484
+ ++++++ P+ + +DN E +YG+ + + DL ++ + S
Sbjct 423 KGIQEVVESLRPNNFFAILAAHSIEKGLDNKEK--FYGIDYGLEDLDSQFIDSLLHIKTS 480
Query 485 PNAFRMPPPLMHIPKASELKILPGLLGLTEPELISEQGGNAGTAVWWQGQGAFALPRIAV 544
+ +P IP + E++ P L P L+ + +W + F +P+ V
Sbjct 481 SELY-LPLANEFIPWSLEVEKQPVTTKLKVPNLVR---NDKFVRLWHKKDDTFWVPKANV 536
Query 545 QLN--GSISKEKADVLSRTQGSLALAAIAEHLQEETVDFQNCGITHSLAFKGTGFHMTFE 602
+N I++ V T +L I + L E + G++ SL+ G +
Sbjct 537 FINFISPIARRSPKVSVST--TLYTRLIEDALGEYSYPASLAGLSFSLSPSTRGIILCIS 594
Query 603 GYTQAQLGKVMAHVASLLSDPSMVEPERFERIKEKQIKLLADPATSMAFEHALEAAAILT 662
G+T +L ++ V +++ D V P+RFE +K + + L D A+ + L+
Sbjct 595 GFTD-KLHVLLEKVVAMMRDLK-VHPQRFEILKNRLEQELKDYDALEAYHRSNHVLTWLS 652
Query 663 RNDAFSRKDVLNAL---QQSNYEDSIAKLSELKNIHVDAFVMGNIDRDQSLAMVEDFLEQ 719
++S ++ A+ Q + D I+ L LK +++ V GN + + ++E +
Sbjct 653 EPHSWSNAELREAIKDVQVGDMSDFISDL--LKQNFLESLVHGNYTEEDAKNLIESAQKL 710
Query 720 AGFTPIAHDDAVASLAM---EQKQTIEATLANPIKGDKDHASLVQFQLGIPSIEDRVNLA 776
P+ A+ E I T+ P K +K+ A + + L I ++D + A
Sbjct 711 IDPKPVFASQLSRKRAIIVPEGGNYIYKTVV-PNKEEKNSA--IMYNLQISQLDDERSGA 767
Query 777 VL---TQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKT----------- 822
+ Q + + LRT+ QLGYI Q L FV+ ++
Sbjct 768 LTRLARQIMKEPTFSILRTKEQLGYIVFTLVRQVTPFINLNIFVQSERSSTYLESRIRAL 827
Query 823 ---HPDEVVKMIDEELTKAKEYL 842
E ++M DE+ +K K L
Sbjct 828 LDQFKSEFLEMSDEDFSKHKSSL 850
> CE06756
Length=845
Score = 244 bits (624), Expect = 5e-64, Method: Compositional matrix adjust.
Identities = 217/850 (25%), Positives = 374/850 (44%), Gaps = 71/850 (8%)
Query 8 SDILKPEADYRDFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLE 67
+ I+K D R+ R +L+NG+ + V P ++++ ++A G L DP ++PGLAHF E
Sbjct 14 NSIVKGPQDERECRGLELTNGLRVLLVSDPTTDKSAVSLAVKAGHLMDPWELPGLAHFCE 73
Query 68 HMLFLGTSKYPEPESYDSFLTQNGGANNAYTDEEKTVFFNKVTDSAFEEALDRFAEFFKS 127
HMLFLGTSKYP + FL+ N G+ NA T+ + T + V ALDRF +FF
Sbjct 74 HMLFLGTSKYPLENEFTKFLSDNAGSYNACTEPDHTYYHFDVKPDQLYGALDRFVQFFLC 133
Query 128 PLFNRQYEEKEVNAIDAEHQKNIPNDDERAWYTIRSLAK-GPMSR-FATGNAETLSTYPK 185
P F + E+EV A+D+EH N+ +D R RSL++ G +R F TGN +TL +
Sbjct 134 PQFTKSATEREVCAVDSEHLSNLNSDYWRILQVDRSLSRPGHDNRKFCTGNKKTLLEDAR 193
Query 186 ANGIDLVDRLRDFHSKYYCGSNMVAVTISPRSLDEQESLIR----EKLEGISAGHADWLG 241
GI+ D L +F+ K+Y + M I SLD ES +R + ++ A W
Sbjct 194 KKGIEPRDALLEFYKKWYSSNIMTCCIIGKESLDVLESYLRTLEFDAIQNKKAESKVWAE 253
Query 242 MVQCPGPMFDTVKPFDHTNSGKFIHLQSFSSQPSLWVAFGLPATLTSYKKQPTSVLTYLL 301
P + K I + + + + F P Y QP + +L+
Sbjct 254 FQYGPDQL------------AKKIDVVPIKDKKLVSIIFPFPDLNNEYLSQPGHYIAHLI 301
Query 302 EYTGEGSLAKRLRLLGLADGISPAVDRNSISTLLG---IKVDLTQKGAAHRGLVLQEIFS 358
+ G GS++ L+ LG A + P + +I+ G + +DL+ +G H ++Q +F+
Sbjct 302 GHKGPGSISSELKRLGWASSLKP--ESKTIAAGFGYFNVTMDLSTEGLEHVDEIIQLMFN 359
Query 359 YINFLRDHGVGHDLVSTMAQQSHIDFHTTQPSSSIMDEAARLAHNLLTYEPY-HVVAGDS 417
YI L+ G + +A+ S I+F + + A ++A N L Y P+ H+++
Sbjct 360 YIGMLQSAGPQQWIHEELAELSAIEFR-FKDREPLTKNAIKVARN-LQYIPFEHILSSRY 417
Query 418 LLIDADPRLTNQLLQAMSPSKAIIAFSDPDFTSKVDNFETDPYYGVQFRVLDL-PQHHAV 476
LL +P +LL ++PS ++ F + N +P YG + +V D+ P+
Sbjct 418 LLTKYNPERIKELLSTLTPSNMLVRVVSKKFKEQEGN-TNEPVYGTEMKVTDISPEKMKK 476
Query 477 AMAVLTASPNAFRMPPPLMHIPKASELKILPGLLGLTEPELISEQGGNAGTAVWWQGQGA 536
L S +A +P +I + K + P LIS+ G + VW++
Sbjct 477 YENALKTSHHALHLPEKNEYIVTKFDQKPRESVKN-EHPRLISDDG---WSRVWFKQDDE 532
Query 537 FALPRIAVQLNGSISKEKADVLSRTQGSLALAAIAEHLQEETVDFQNCGITHSLAFKGTG 596
+ +P+ +L + + + SL L + + L EET + G+ L G
Sbjct 533 YNMPKQETKLAFTTPIVAQNPIMSLISSLWLWCLNDTLTEETYNAAIAGLKFQLESGHNG 592
Query 597 FH----------------MTFEGYTQAQLGKVMAHVASLLSDPSMVEPERFERIKEKQIK 640
H + GY + Q + H+ +++ ++ RF+ + E +
Sbjct 593 VHEQAGNWLDPERHASITLHVYGYDEKQ-PLFVKHLTKCMTN-FKIDRTRFDVVFESLKR 650
Query 641 LLADPATSMAFEHALEAAAILTRNDAFSRKDVLNALQQSNYEDSIAKLSEL-KNIHVDAF 699
L + A S + + +L +S++ +L + ED EL + H++ F
Sbjct 651 SLTNHAFSQPYMLSKYFNELLVVEKVWSKEQLLAVCDSATLEDVQGFSKELFQAFHLELF 710
Query 700 VMGNIDRDQSLAMVEDFLEQAGFTPIAHDDAVASLAMEQKQTIEATLANPIKGD------ 753
V GN +++ + + ++ A ++ E E L N GD
Sbjct 711 VHGNSTEKKAIQLSNELMD---ILKSAAPNSRLLYRNEHNPRREFQLNN---GDEYIYRH 764
Query 754 --KDH---ASLVQFQLGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLG---YIAGAKES 805
K H V F+ G+ + D +++Q + + + +LRT+ LG Y K S
Sbjct 765 LQKTHDAGCVEVTFKFGVQNTYDNALAGLISQLIRQPAFSTLRTKESLGKNSYPHLIKYS 824
Query 806 QAASTALLQC 815
+ LL+C
Sbjct 825 KYLERKLLRC 834
> At1g06900
Length=1023
Score = 239 bits (611), Expect = 1e-62, Method: Compositional matrix adjust.
Identities = 200/787 (25%), Positives = 360/787 (45%), Gaps = 53/787 (6%)
Query 38 RSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPEPESYDSFLTQNGGANNAY 97
++ +A A+ + GS DP + GLAHFLEHMLF+G++++P+ YDS+L+++GG++NAY
Sbjct 103 QTKKAAAAMCVSMGSFLDPPEAQGLAHFLEHMLFMGSTEFPDENEYDSYLSKHGGSSNAY 162
Query 98 TDEEKTVFFNKVTDSAFEEALDRFAEFFKSPLFNRQYEEKEVNAIDAEHQKNIPNDDER- 156
T+ E T + +V + AL RF++FF +PL + E+EV A+D+E + + ND R
Sbjct 163 TEMEHTCYHFEVKREFLQGALKRFSQFFVAPLMKTEAMEREVLAVDSEFNQALQNDACRL 222
Query 157 AWYTIRSLAKG-PMSRFATGNAETLSTYPKANGIDLVDRLRDFHSKYYCGSNMVAVTISP 215
+ AKG P +RFA G + + NG+DL + + + +YY G M V I
Sbjct 223 QQLQCYTSAKGHPFNRFAWGKDTAVLSGAMENGVDLRECIVKLYKEYYHGGLMKLVVIGG 282
Query 216 RSLDEQESLIREKLEGISAGHADWLGMVQCPGPMFDTVKPFDHTNSGKFIHLQSFSSQPS 275
SLD ES + E + G + ++ GP++ GK L++
Sbjct 283 ESLDMLESWVVELFGDVKNG-SKIRPTLEAEGPIW---------KGGKLYRLEAVKDVHI 332
Query 276 LWVAFGLPATLTSYKKQPTSVLTYLLEYTGEGSLAKRLRLLGLADGISPAV-----DRNS 330
L + + LP ++Y K+P L +LL + G GSL L+ G A +S V +R+S
Sbjct 333 LDLTWTLPPLRSAYVKKPEDYLAHLLGHEGRGSLHSFLKAKGWATSLSAGVGDDGINRSS 392
Query 331 ISTLLGIKVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHDLVSTMAQQSHIDFH--TTQ 388
++ + G+ + LT G ++ I+ Y+ LRD + + ++DF Q
Sbjct 393 LAYVFGMSIHLTDSGLEKIYDIIGYIYQYLKLLRDVSPQEWIFKELQDIGNMDFRFAEEQ 452
Query 389 PSSSIMDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQAMSPSKAIIAFSDPDF 448
P+ D AA L+ N+L Y HV+ GD + DP+L L+ +P I
Sbjct 453 PAD---DYAAELSENMLAYPVEHVIYGDYVYQTWDPKLIEDLMGFFTPQNMRIDVVSKSI 509
Query 449 TSKVDNFETDPYYGVQFRVLDLPQHHAVAMAVLTASPNAFRMPPPLMHIPKASELKILPG 508
S + F+ +P++G + D+P + + + N+ +P IP ++ +
Sbjct 510 KS--EEFQQEPWFGSSYIEEDVPLSLMESWSNPSEVDNSLHLPSKNQFIPCDFSIRAINS 567
Query 509 LL---GLTEPELISEQGGNAGTAVWWQGQGAFALPRIAVQLNGSISKEKADVLSRTQGSL 565
+ + P I ++ W++ F +PR ++ A V + L
Sbjct 568 DVDPKSQSPPRCIIDE---PFMKFWYKLDETFKVPRANTYFRINLKGAYASVKNCLLTEL 624
Query 566 ALAAIAEHLQEETVDFQNCGITHSLAFKGTGFHMTFEGYTQ---AQLGKVMAHVASLLSD 622
+ + + L E + +Q + SL+ G + G+ + A L K++A S +
Sbjct 625 YINLLKDELNE--IIYQATKLETSLSMYGDKLELKVYGFNEKIPALLSKILAIAKSFM-- 680
Query 623 PSMVEPERFERIKEKQIKLLADPATSMA-FEHALEAAAILTRNDAFSRKDVLNALQQSNY 681
P++ ERF+ IKE + + T+M H+ L + + L+ L +
Sbjct 681 PNL---ERFKVIKENMERGFRN--TNMKPLNHSTYLRLQLLCKRIYDSDEKLSVLNDLSL 735
Query 682 EDSIAKLSELKN-IHVDAFVMGNIDRDQSLAMVEDFLEQAGFTPI----AHDDAVASLAM 736
+D + + EL++ I ++A GN+ D+++ + F + P+ H + + M
Sbjct 736 DDLNSFIPELRSQIFIEALCHGNLSEDEAVNISNIFKDSLTVEPLPSKCRHGEQITCFPM 795
Query 737 EQKQTIEATLANPIKGDKDHASLVQFQLGIPSIEDRVNLAVLTQF---LNRRIYDSLRTE 793
K + + N K + + + +Q+ + AVL F + +++ LRT+
Sbjct 796 GAKLVRDVNVKN--KSETNSVVELYYQIEPEEAQSTRTKAVLDLFHEIIEEPLFNQLRTK 853
Query 794 AQLGYIA 800
QLGY+
Sbjct 854 EQLGYVV 860
> Hs4505453
Length=1219
Score = 239 bits (610), Expect = 2e-62, Method: Compositional matrix adjust.
Identities = 209/778 (26%), Positives = 352/778 (45%), Gaps = 55/778 (7%)
Query 45 AVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPEPESYDSFLTQNGGANNAYTDEEKTV 104
A+ GS DP+D+PGLAHFLEHM+F+G+ KYP+ +D+FL ++GG++NA TD E+TV
Sbjct 282 ALCVGVGSFADPDDLPGLAHFLEHMVFMGSLKYPDENGFDAFLKKHGGSDNASTDCERTV 341
Query 105 FFNKVTDSAFEEALDRFAEFFKSPLFNRQYEEKEVNAIDAEHQKNIPNDDERAWYTIRSL 164
F V F+EALDR+A+FF PL R ++EV A+D+E+Q P+D R SL
Sbjct 342 FQFDVQRKYFKEALDRWAQFFIHPLMIRDAIDREVEAVDSEYQLARPSDANRKEMLFGSL 401
Query 165 AK--GPMSRFATGNAETLSTYPKANGIDLVDRLRDFHSKYYCGSNMVAVTISPRSLDEQE 222
A+ PM +F GNAETL P+ N ID RLR+F +YY M V S +LD E
Sbjct 402 ARPGHPMGKFFWGNAETLKHEPRKNNIDTHARLREFWMRYYSSHYMTLVVQSKETLDTLE 461
Query 223 SLIREKLEGISAGHADWLGMVQCPGPMFDTVK-PFDHTNSGKFIHLQSFSSQPSLWVAFG 281
+ E I G+ P P F + PFD K + +L + +
Sbjct 462 KWVTEIFSQIPNN-----GL---PRPNFGHLTDPFDTPAFNKLYRVVPIRKIHALTITWA 513
Query 282 LPATLTSYKKQPTSVLTYLLEYTGEGS----LAKRLRLLGLADGIS-PAVDRNSISTLLG 336
LP Y+ +P +++L+ + G+GS L K+ L L G ++NS ++
Sbjct 514 LPPQQQHYRVKPLHYISWLVGHEGKGSILSFLRKKCWALALFGGNGETGFEQNSTYSVFS 573
Query 337 IKVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHDLVSTMAQQSHIDFHTTQPSSSIMDE 396
I + LT +G H V +F Y+ L+ G + + + +FH Q + ++
Sbjct 574 ISITLTDEGYEHFYEVAYTVFLYLKMLQKLGPEKRIFEEIRKIEDNEFH-YQEQTDPVEY 632
Query 397 AARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQAMSPSKAIIAFSDPDFTSKVDNFE 456
+ N+ Y ++ GD LL + P + + L + P KA + K D E
Sbjct 633 VENMCENMQLYPLQDILTGDQLLFEYKPEVIGEALNQLVPQKANLVLLSGANEGKCDLKE 692
Query 457 TDPYYGVQFRVLDLPQHHAVAMAVLTASPNAFRMPPPLMHIPKASELKILPGLLGLT--- 513
++G Q+ + D+ + A L S F + P L H+P +E K + L
Sbjct 693 K--WFGTQYSIEDIEN----SWAELWNS--NFELNPDL-HLP--AENKYIATDFTLKAFD 741
Query 514 --EPELISEQGGNAGTAVWWQGQGAFALPRIAVQ---LNGSISKEKADVLSRTQGSLALA 568
E E + +W++ F +P+ ++ ++ I K A+V+ + +
Sbjct 742 CPETEYPVKIVNTPQGCLWYKKDNKFKIPKAYIRFHLISPLIQKSAANVV---LFDIFVN 798
Query 569 AIAEHLQEETVDFQNCGITHSLAFKGTGFHMTFEGYTQA-----QLGKVMAHVASLLSDP 623
+ +L E + + + LA G + +G+ QL ++ ++A S P
Sbjct 799 ILTHNLAEPAYEADVAQLEYKLAAGEHGLIIRVKGFNHKLPLLFQL--IIDYLAEFNSTP 856
Query 624 SMVEPERFERIKEKQIKLLADPATSMAFEHALEAAAILTRNDAFSRKDVLNALQQS-NYE 682
+ V E++K+ +L P T A + ++ +S D AL + E
Sbjct 857 A-VFTMITEQLKKTYFNILIKPETL-----AKDVRLLILEYARWSMIDKYQALMDGLSLE 910
Query 683 DSIAKLSELKN-IHVDAFVMGNIDRDQSLAMVEDFLEQAGFTPIAHDDAVASLAMEQKQT 741
++ + E K+ + V+ V GN+ +S+ ++ +++ F P+ + V +E
Sbjct 911 SLLSFVKEFKSQLFVEGLVQGNVTSTESMDFLKYVVDKLNFKPLEQEMPVQFQVVELPSG 970
Query 742 IEATLANPI-KGDKDHASLVQFQLGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGY 798
+ KGD + V +Q G S+ + + +L + +D LRT+ LGY
Sbjct 971 HHLCKVKALNKGDANSEVTVYYQSGTRSLREYTLMELLVMHMEEPCFDFLRTKQTLGY 1028
> CE10402
Length=1067
Score = 233 bits (595), Expect = 1e-60, Method: Compositional matrix adjust.
Identities = 215/878 (24%), Positives = 375/878 (42%), Gaps = 79/878 (8%)
Query 10 ILKPEADYRDFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHM 69
I+K D R++R +L+NG+ + V P ++++ A+ G L DP ++PGLAHF EHM
Sbjct 75 IVKGAQDAREYRGLELTNGIRVLLVSDPTTDKSAAALDVKVGHLMDPWELPGLAHFCEHM 134
Query 70 LFLGTSKYPEPESYDSFLTQNGGANNAYTDEEKTVFFNKVTDSAFEEALDRFAEFFKSPL 129
LFLGT+KYP Y FL + G++NAYT + T + V ALDRF +FF SP
Sbjct 135 LFLGTAKYPSENEYSKFLAAHAGSSNAYTSSDHTNYHFDVKPDQLPGALDRFVQFFLSPQ 194
Query 130 FNRQYEEKEVNAIDAEHQKNIPNDDERAWYTIRSLAK--GPMSRFATGNAETLSTYPKAN 187
F E+EV A+D+EH N+ ND R RS +K +F TGN +TL +
Sbjct 195 FTESATEREVCAVDSEHSNNLNNDLWRFLQVDRSRSKPGHDYGKFGTGNKQTLLEDARKK 254
Query 188 GIDLVDRLRDFHSKYYCGSNMVAVTISPRSLDEQESLIREKLEGISAGHADWLGMVQCPG 247
GI+ D L FH K+Y M + L+ ES +LG ++
Sbjct 255 GIEPRDALLQFHKKWYSSDIMTCCIVGKEPLNVLES---------------YLGTLE--- 296
Query 248 PMFDTVK------------PFDHTNSGKFIHLQSFSSQPSLWVAFGLPATLTSYKKQPTS 295
FD ++ P+ K I + + ++F P + QP
Sbjct 297 --FDAIENKKVERKVWEEFPYGPDQLAKRIDVVPIKDTRLVSISFPFPDLNGEFLSQPGH 354
Query 296 VLTYLLEYTGEGSLAKRLRLLGLADGISPAVDRNSISTLLG---IKVDLTQKGAAHRGLV 352
+++L+ + G GSL L+ LG + D ++ + G + +DL+ +G H +
Sbjct 355 YISHLIGHEGPGSLLSELKRLGWVSSLQS--DSHTQAAGFGVYNVTMDLSTEGLEHVDEI 412
Query 353 LQEIFSYINFLRDHGVGHDLVSTMAQQSHIDFH---TTQPSSSIMDEAARLAHNLLTYEP 409
+Q +F+YI L+ G + +A+ S + F QP + ++ AA L Y P
Sbjct 413 IQLMFNYIGMLQSAGPKQWVHDELAELSAVKFRFKDKEQPMTMAINVAAS-----LQYIP 467
Query 410 Y-HVVAGDSLLIDADPRLTNQLLQAMSPSKAIIAFSDPDFTSKVDNFETDPYYGVQFRVL 468
+ H+++ LL +P +LL +SP+ + F + N +P YG + +V
Sbjct 468 FEHILSSRYLLTKYEPERIKELLSMLSPANMQVRVVSQKFKGQEGN-TNEPVYGTEMKVT 526
Query 469 DL-PQHHAVAMAVLTASPNAFRMPPPLMHIPKASELKILPGLLGLTEPELISEQGGNAGT 527
D+ P+ L S +A +P +I + K + P LIS+ G +
Sbjct 527 DISPETMKKYENALKTSHHALHLPEKNEYIATNFDQKPRESVKN-EHPRLISDDG---WS 582
Query 528 AVWWQGQGAFALPRIAVQLNGSISKEKADVLSRTQGSLALAAIAEHLQEETVDFQNCGIT 587
VW++ + +P+ +L + + SL L +++ L EET + G+
Sbjct 583 RVWFKQDDEYNMPKQETKLALTTPMVAQNPRMSLLSSLWLWCLSDTLAEETYNADLAGLK 642
Query 588 HSLAFKGTGFHMTFE----------------GYTQAQLGKVMAHVASLLSDPSMVEPERF 631
L G M GY + Q H+A+ +++ ++ RF
Sbjct 643 CQLESSPFGVQMRVSNRREAERHASLTLHVYGYDEKQ-ALFAKHLANRMTN-FKIDKTRF 700
Query 632 ERIKEKQIKLLADPATSMAFEHALEAAAILTRNDAFSRKDVLNALQQSNYEDSIAKLSE- 690
+ + E + L + A S + +L + +S++ +L ED E
Sbjct 701 DVLFESLKRALTNHAFSQPYLLTQHYNQLLIVDKVWSKEQLLAVCDSVTLEDVQGFAKEM 760
Query 691 LKNIHVDAFVMGNIDRDQSLAMVEDFLE-----QAGFTPIAHDDAVASLAMEQKQTIEAT 745
L+ H++ FV GN +++ + ++ ++ P+ ++ ++ E
Sbjct 761 LQAFHMELFVHGNSTEKEAIQLSKELMDVLKSAAPNSRPLYRNEHNPRRELQLNNGDEYV 820
Query 746 LANPIKGDKDHASLVQFQLGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKES 805
+ K V +Q+G+ + D + ++ Q + +++LRT LGYI
Sbjct 821 YRHLQKTHDVGCVEVTYQIGVQNTYDNAVVGLIDQLIREPAFNTLRTNEALGYIVWTGSR 880
Query 806 QAASTALLQCFVEGAKTHPDEVVKMIDEELTKAKEYLA 843
T L V+G K+ D V++ I+ L ++ +A
Sbjct 881 LNCGTVALNVIVQGPKS-VDHVLERIEVFLESVRKEIA 917
> CE06755
Length=980
Score = 230 bits (586), Expect = 1e-59, Method: Compositional matrix adjust.
Identities = 217/846 (25%), Positives = 363/846 (42%), Gaps = 83/846 (9%)
Query 10 ILKPEADYRDFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHM 69
I+K D R++R +L+NG+ + V P ++++ A+ G L DP ++PGLAHF EHM
Sbjct 16 IVKGAQDDREYRGLELTNGIRVLLVSDPTTDKSAAALDVKVGHLMDPWELPGLAHFCEHM 75
Query 70 LFLGTSKYPEPESYDSFLTQNGGANNAYTDEEKTVFFNKVTDSAFEEALDRFAEFFKSPL 129
LFLGT+KYP Y FLT N G NA T + T + V ALDRF +FF SP
Sbjct 76 LFLGTAKYPTENEYSKFLTDNAGHRNAVTASDHTNYHFDVKPDQLRGALDRFVQFFLSPQ 135
Query 130 FNRQYEEKEVNAIDAEHQKNIPNDDERAWYTIRSLAK--GPMSRFATGNAETLSTYPKAN 187
F E+EV A+D+EH N+ ND R RSL+K ++F TGN +TL +
Sbjct 136 FTESATEREVCAVDSEHSNNLNNDLWRLSQVDRSLSKPGHDYAKFGTGNKKTLLEEARKK 195
Query 188 GIDLVDRLRDFHSKYYCGSNMVAVTISPRSLDEQESLIREKLEGISAGHADWLGMVQCPG 247
G++ D L F+ K+Y + M I SLD +S ++ LE
Sbjct 196 GVEPRDALLQFYKKWYSSNIMTCCIIGKESLDVLQSHLK-TLE----------------- 237
Query 248 PMFDTVKPFDHTNSGKFIHLQSFSSQPSLWVAFGLPATLTSYKKQPTSVLTYLLEYTGEG 307
FDT++ K + + ++ P +G P L K+ +L+ + G G
Sbjct 238 --FDTIE-------NKKVERKVWNENP-----YG-PEQLG--KRIDRKFFAHLIRHKGPG 280
Query 308 SLAKRLRLLGLADGISPAVDRNSIST---LLGIKVDLTQKGAAHRGLVLQEIFSYINFLR 364
SL L+ LG + + D N+I+ +L + +DL+ G + ++Q + +YI L+
Sbjct 281 SLLVELKRLGWVNSLKS--DSNTIAAGFGILNVTMDLSTGGLENVDEIIQLMLNYIGMLK 338
Query 365 DHGVGHDLVSTMAQQSHIDFH---TTQPSSSIMDEAARLAHNLLTYEPY-HVVAGDSLLI 420
G + +A S + F QP ++ AA L Y P H+++ LL
Sbjct 339 SFGPQQWIHDELADLSDVKFRFKDKEQPMKMAINIAASLQ-----YIPIEHILSSRYLLT 393
Query 421 DADPRLTNQLLQAMSPSKAIIAFSDPDFTSKVDNFETDPYYGVQFRVLDL-PQHHAVAMA 479
+P +LL ++PS ++ F + N +P YG + +V D+ P+
Sbjct 394 KYEPERIKELLSTLTPSNMLVRVVSQKFKEQEGN-TNEPVYGTEMKVTDISPEKMKKYEN 452
Query 480 VLTASPNAFRMPPPLMHIPKASELKILPGLLGLTE-PELISEQGGNAGTAVWWQGQGAFA 538
L S +A +P +I A+ P E P+LIS+ G + VW++ +
Sbjct 453 ALKTSHHALHLPEKNEYI--ATNFGQKPRESVKNEHPKLISDDG---WSRVWFKQDDEYN 507
Query 539 LPRIAVQL---------NGSISKEKA-------DVLSRTQGSLALAAIAEHLQEETVDFQ 582
+P+ + N IS + D+LS + ALA + + Q
Sbjct 508 MPKQETKFALTTPIVSQNPRISLISSLWLWCFCDILSEETYNAALAGLGCQFELSPFGVQ 567
Query 583 NCGITHSLAFKGTGFHMTFEGYTQAQLGKVMAHVASLLSDPSMVEPERFERIKEKQIKLL 642
A + + GY + Q + H+ S + + ++ RFE + E + L
Sbjct 568 KQSTDGREAERHASLTLHVYGYDEKQ-PLFVKHLTSCMIN-FKIDRTRFEVLFESLKRTL 625
Query 643 ADPATSMAFEHALEAAAILTRNDAFSRKDVLNALQQSNYEDSIAKLSE-LKNIHVDAFVM 701
+ A S + +L + +S++ +L E+ E L+ H++ FV
Sbjct 626 TNHAFSQPYLLTQHYNQLLIVDKVWSKEQLLAVCDSVTLENVQGFAREMLQAFHMELFVH 685
Query 702 GNIDRDQSLAMVE---DFLEQAG--FTPIAHDDAVASLAMEQKQTIEATLANPIKGDKDH 756
GN +++ + + D L+ A P+ ++ + E + K
Sbjct 686 GNSTEKEAIQLSKELMDILKSAAPNSRPLYRNEHNPRREFQLNNGDEYIYRHLQKTHDAG 745
Query 757 ASLVQFQLGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCF 816
V +Q+G+ + D + ++ Q + ++D+LRT LGYI L F
Sbjct 746 CVEVTYQIGVQNKYDNAVVGLIDQLIKEPVFDTLRTNEALGYIVWTGCRFNCGAVALNIF 805
Query 817 VEGAKT 822
V+G K+
Sbjct 806 VQGPKS 811
> CE01818
Length=745
Score = 187 bits (474), Expect = 1e-46, Method: Compositional matrix adjust.
Identities = 129/411 (31%), Positives = 196/411 (47%), Gaps = 43/411 (10%)
Query 10 ILKPEADYRDFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHM 69
I+K D + +R +L+NG+ + V ++ + A+ G L DP ++PGLAHF EHM
Sbjct 17 IVKGSQDTKKYRGLELTNGLRVLLVSDSKTRVSAVALDVKVGHLMDPWELPGLAHFCEHM 76
Query 70 LFLGTSKYPEPESYDSFLTQNGGANNAYTDEEKTVFFNKVTDSAFEEALDRFAEFFKSPL 129
LFLGT+KYP Y +L N G +NAYTD + T + +V ALDRFA+FF P
Sbjct 77 LFLGTAKYPSEREYFKYLAANNGDSNAYTDTDHTNYSFEVRSEKLYGALDRFAQFFLDPQ 136
Query 130 FNRQYEEKEVNAIDAEHQKNIPNDDERAWYTIRSLAK--GPMSRFATGNAETLSTYPKAN 187
F E+EV A++ E+ + D R RSL+K S+FA GN +TL P+
Sbjct 137 FTESATEREVCAVNCEYLDKVNEDFWRCLQVERSLSKPGHDYSKFAIGNKKTLLEDPRTK 196
Query 188 GIDLVDRLRDFHSKYYCGSNMVAVTISPRSLDEQESLIREKLEGISAGHADWLGMVQCPG 247
GI+ D L DF+ +Y M + SLD ES +LG +
Sbjct 197 GIEPRDVLLDFYKNWYSSDIMTCCIVGKESLDVLES---------------YLGSFK--- 238
Query 248 PMFDTVK------------PFDHTNSGKFIHLQSFSSQPSLWVAFGLPATLTSYKKQPTS 295
FD +K PF K I + + + + F P + QP
Sbjct 239 --FDAIKNTRKERKIWKDSPFGPDQLAKRIEIVPIQNTGQVSIKFPFPDLNGEFLSQPGD 296
Query 296 VLTYLLEYTGEGSLAKRLRLLGLADGISPAVDRNSISTLLG---IKVDLTQKGAAHRGLV 352
+ +L+ + G GSL L+ LG IS D ++I++ G + +DL+ +G H V
Sbjct 297 YIAHLIGHEGPGSLLSELKRLGWV--ISLEADNHTIASGFGVFSVTMDLSTEGLEHVDDV 354
Query 353 LQEIFSYINFLRDHGVGHDLVSTMAQQSHIDFHTTQPSSSIMDEA--ARLA 401
+Q +F++I FL+ G + +A+ + +DF +I++E ARLA
Sbjct 355 IQLVFNFIGFLKSSGPQKWIHDELAELNAVDFRFD--DDTIIEETYNARLA 403
Score = 35.8 bits (81), Expect = 0.46, Method: Compositional matrix adjust.
Identities = 55/273 (20%), Positives = 104/273 (38%), Gaps = 13/273 (4%)
Query 576 EETVDFQNCGITHSLAFKGTGFHMTFEGYTQAQ--LGKVMAHVASLLSDPSMVEPERFER 633
EET + + G+ +G + GY + Q K + + + + FE
Sbjct 395 EETYNARLAGLECQFESSSSGVQIRVFGYDEKQSLFAKHLVNRMANFQVNRLCFDISFES 454
Query 634 IKEKQIKLLADPATSMAFEHALEAAAILTRNDAFSRKDVLNALQQSNYEDSIA-KLSELK 692
+K + L + A S + + +L ++ +S++ +L ED + L+
Sbjct 455 LK----RTLTNHAFSQPHDLSAHFIDLLVVDNIWSKEQLLAVCDSVTLEDVHGFAIKMLQ 510
Query 693 NIHVDAFVMGNIDRDQSLAM---VEDFLEQAG--FTPIAHDDAVASLAMEQKQTIEATLA 747
H++ FV GN +L + + D L+ P+ D+ ++ E
Sbjct 511 AFHMELFVHGNSTEKDTLQLSKELSDILKSVAPNSRPLKRDEHNPHRELQLINGHEHVYR 570
Query 748 NPIKGDKDHASLVQFQLGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQA 807
+ K V FQ+G+ S + +L + + Y LRT LGY +
Sbjct 571 HFQKTHDVGCVEVAFQIGVQSTYNNSVNKLLNELIKNPAYTILRTNEALGYNVSTESRLN 630
Query 808 ASTALLQCFVEGAKTHPDEVVKMIDEELTKAKE 840
L V+G ++ D V++ I+ L A+E
Sbjct 631 DGNVYLHVIVQGPES-ADHVLERIEVFLESARE 662
> 7292708
Length=1077
Score = 176 bits (447), Expect = 2e-43, Method: Compositional matrix adjust.
Identities = 193/805 (23%), Positives = 347/805 (43%), Gaps = 88/805 (10%)
Query 42 AGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPEPESYDSFLTQNGGANNAYTDEE 101
A A+ + GS +P GLAHFLEHM+F+G+ KYP+ +D+ + + GG NA TD E
Sbjct 94 AACALLIDYGSFAEPTKYQGLAHFLEHMIFMGSEKYPKENIFDAHIKKCGGFANANTDCE 153
Query 102 KTVFFNKVTDSAFEEALDRFAEFFKSPLFNRQYEEKEVNAIDAEHQKNIPNDDERAWYTI 161
T+F+ +V + + +LD F K+PL ++ ++E +A+D+E Q+ + +D+ R +
Sbjct 154 DTLFYFEVAEKHLDSSLDYFTALMKAPLMKQEAMQRERSAVDSEFQQILQDDETRRDQLL 213
Query 162 RSLA-KG-PMSRFATGNAETLSTYPKANGIDLVDRLRDFHSKYYCGSNMVAVTISPR-SL 218
SLA KG P FA GN ++L + +L L + ++Y G+N + V + R +
Sbjct 214 ASLATKGFPHGTFAWGNMKSLKE--NVDDAELHKILHEIRKEHY-GANRMYVCLQARLPI 270
Query 219 DEQESLIREKLEGISAGHADWLGMVQCPGPMFDTVKPFDHTNSGKF-IHLQSFSSQP--- 274
DE ESL+ GI V+ P + F++ ++ K H Q F +P
Sbjct 271 DELESLVVRHFSGIPHNE------VKAPD-----LSSFNYKDAFKAEFHEQVFFVKPVEN 319
Query 275 --SLWVAFGLPATLTSYKKQPTSVLTYLLEYTGEGSLA----KRLRLLGLADGISP-AVD 327
L + + LP Y+ +P L+YLL Y G GSL +RL L L GI D
Sbjct 320 ETKLELTWVLPNVRQYYRSKPDQFLSYLLGYEGRGSLCAYLRRRLWALQLIAGIDENGFD 379
Query 328 RNSISTLLGIKVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHDLVSTMA--QQSHIDFH 385
NS+ +L I + LT +G + VL F+Y+ + G D+ +++ FH
Sbjct 380 MNSMYSLFNICIYLTDEGFKNLDEVLAATFAYVKLFANCGSMKDVYEEQQRNEETGFRFH 439
Query 386 TTQPSSSIMDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQAMSPSK---AIIA 442
+P+ D L NL + P ++ G L + + +L+ ++ K + +
Sbjct 440 AQRPA---FDNVQELVLNLKYFPPKDILTGKELYYEYNEEHLKELISHLNEMKFNLMVTS 496
Query 443 FSDPDFTSKVDNFETDPYYGVQFRVLDLPQHHAVAMAVLTASPNAFRMPPPLMHIPKASE 502
D S D +T+ ++G ++ + +P+ P F +P ++ +
Sbjct 497 RRKYDDISAYD--KTEEWFGTEYATIPMPEKWRKLWEDSVPLPELF-LPESNKYV--TDD 551
Query 503 LKILPGLLGLTE----PELISEQGGNAGTAVWWQGQGAFALPRIAVQLNGSISKEKADVL 558
+ +G E P+L+ + +W++ F LP + ++ +
Sbjct 552 FTLHWHSMGRPEVPDSPKLLIKTD---TCELWFRQDDKFDLPEAHMAFYFISPMQRQNAK 608
Query 559 SRTQGSLALAAIAEHLQEETVDFQNCGITHSLAFKGTGFHMTFEGYTQAQLGKVMAHVAS 618
+ SL + H+ EE + G+++SL+ G + GY + V A
Sbjct 609 NDAMCSLYEEMVRFHVCEELYPAISAGLSYSLSTIEKGLLLKVCGYNEKLHLIVEAIAEG 668
Query 619 LLSDPSMVEPERFERIKEKQIKLLADPATSMAFEHALEAAAILTRNDAFSRKDVLNALQQ 678
+L+ ++ + Q K AF +AL L R+ + + L
Sbjct 669 MLNVAETLDENMLSAFVKNQRK---------AFFNALIKPKALNRDIRLCVLERIRWLMI 719
Query 679 SNYED-SIAKLSEL--------KNIHVDAFVMGNIDRDQSLAMVEDFLEQAGFTPIAHDD 729
+ Y+ S L ++ K +++ + + GN + + ++ L + I
Sbjct 720 NKYKCLSSVILEDMREFAHQFPKELYIQSLIQGNYTEESAHNVMNSLLSRLNCKQIR--- 776
Query 730 AVASLAMEQKQTIE-ATLANPIKGD--KDHASLVQ---------FQLGIPSIEDRVNLAV 777
E+ + +E T+ P+ + HA VQ +Q+G ++ L +
Sbjct 777 -------ERGRFLEDITVKLPVGTSIIRCHALNVQDTNTVITNFYQIGPNTVRVESILDL 829
Query 778 LTQFLNRRIYDSLRTEAQLGYIAGA 802
L F++ ++D LRT+ QLGY GA
Sbjct 830 LMMFVDEPLFDQLRTKEQLGYHVGA 854
> ECU06g0750
Length=882
Score = 167 bits (423), Expect = 1e-40, Method: Compositional matrix adjust.
Identities = 116/392 (29%), Positives = 194/392 (49%), Gaps = 16/392 (4%)
Query 20 FRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPE 79
+ + ++ NGM A+ + P ++ AV+ GS DP D GLAHFLEHMLF+GT KYP
Sbjct 27 YEYVEIPNGMRALIMSDPGLDKCSCAVSVRVGSFDDPADAQGLAHFLEHMLFMGTEKYPV 86
Query 80 PESYDSFLTQNGGANNAYTDEEKTVFFNKVTDSAFEEALDRFAEFFKSPLFNRQYEEKEV 139
+ FL++N G NA T E TV++ V AFEEA+D FA+FFKSPL R E+EV
Sbjct 87 EDGLSYFLSKNNGGYNATTYGEATVYYFDVRPEAFEEAVDMFADFFKSPLLKRDSVEREV 146
Query 140 NAIDAEHQKNIPNDDERAWYTIRSLAKGP--MSRFATGNAETLSTYPKANGIDLVDRLRD 197
+A+++E + ND R W ++ K +S+F+TGN +TL + +GI + +++
Sbjct 147 SAVNSEFCNGLNNDGWRTWRMMKKCCKKEQALSKFSTGNYDTL----RRDGI--WEEMKE 200
Query 198 FHSKYYCGSNMVAVTISPRSLDEQESLIREKLEGISAGHA-DWLGMVQCPGPMFDTVKPF 256
F S+ Y M V +S E + L+ K EG+ GH D + + F
Sbjct 201 FWSRKYSSDKMCVVIYGNKSPGELKELL-GKFEGVPKGHGEDSCDKKDRKCRLDEGWSVF 259
Query 257 DHTNSGKFIHLQSFSSQPSLWVAFGLPATLTSYKKQPTS-VLTYLLEYTGEGSLAKRLRL 315
D + ++I +Q + S+ V + + +K P VL +L +G A +++
Sbjct 260 DEEYTNRWIRIQPIADTRSIMVMMTVESGYKMFKNNPYEYVLNMILRNDSKG-FACKVKD 318
Query 316 LGLADGISPAVDRNSISTLLGIKVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHDLVST 375
+GL + + + +++ I + L+ G + E+ + +L+ V D +
Sbjct 319 MGLVLEVEGDLCDYTDYSVMIITMHLSAAGNKRPKEAVLEL---VKYLKTMPVCLDEYAE 375
Query 376 MAQQSHIDFHTTQPSSSIMDEAARLAHNLLTY 407
+ ++S F + M + R+A N+ Y
Sbjct 376 LQKRSRYLFKYDEKDDP-MYQTRRVAVNMQFY 406
> YPR122w
Length=1208
Score = 133 bits (334), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 77/229 (33%), Positives = 119/229 (51%), Gaps = 9/229 (3%)
Query 18 RDFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLF-LGTSK 76
R + +L NG+ A+ + P + ++ TGS DP+D+ GLAH EHM+ G+ K
Sbjct 22 RTHKVCKLPNGILALIISDPTDTSSSCSLTVCTGSHNDPKDIAGLAHLCEHMILSAGSKK 81
Query 77 YPEPESYDSFLTQNGGANNAYTDEEKTVFFNKVTDS------AFEEALDRFAEFFKSPLF 130
YP+P + + + +N G+ NA+T E+T F+ ++ ++ FE LD FA FFK PLF
Sbjct 82 YPDPGLFHTLIAKNNGSQNAFTTGEQTTFYFELPNTQNNGEFTFESILDVFASFFKEPLF 141
Query 131 NRQYEEKEVNAIDAEHQKNIPNDDERAWYTIRSLAK--GPMSRFATGNAETLSTYPKANG 188
N KE+ AI +EH+ NI + + ++ R LA P SRF+TGN +LS+ P+
Sbjct 142 NPLLISKEIYAIQSEHEGNISSTTKIFYHAARILANPDHPFSRFSTGNIHSLSSIPQLKK 201
Query 189 IDLVDRLRDFHSKYYCGSNMVAVTISPRSLDEQESLIREKLEGISAGHA 237
I L L + + G N+ P+S++ L K I A
Sbjct 202 IKLKSSLNTYFENNFFGENITLCIRGPQSVNILTKLALSKFGDIKPKSA 250
> 7296373
Length=908
Score = 123 bits (309), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 105/384 (27%), Positives = 174/384 (45%), Gaps = 42/384 (10%)
Query 12 KPEADYRDFRHYQLSNGMHAIAVH---------HPRSNE-------------AGFAVAAN 49
K + D + +R LSNG+ A+ + H S E A AV
Sbjct 42 KSDGDSKLYRALTLSNGLRAMLISDSYIDEPSIHRASRESLNSSTENFNGKLAACAVLVG 101
Query 50 TGSLYDPEDVPGLAHFLEHMLFLGTSKYPEPESYDSFLTQNGGANNAYTDEEKTVFFNKV 109
GS +P+ GLAHF+EHM+F+G+ K+P +DSF+T++GG +NA+T+ E+T F+ ++
Sbjct 102 VGSFSEPQQYQGLAHFVEHMIFMGSEKFPVENEFDSFVTKSGGFSNAHTENEETCFYFEL 161
Query 110 TDSAFEEALDRFAEFFKSPLFNRQYEEKEVNAIDAEHQKNIPNDDERAWYTIRSLAKG-- 167
+ + +D F K+PL +E +A+ +E ++ D+ R + SLA
Sbjct 162 DQTHLDRGMDLFMNLMKAPLMLPDAMSRERSAVQSEFEQTHMRDEVRRDQILASLASEGY 221
Query 168 PMSRFATGNAETLSTYPKANGID---LVDRLRDFHSKYYCGSNMVAVTISPRSLDEQESL 224
P F+ GN +TL G+D L L F+ +Y + MV + SLDE E L
Sbjct 222 PHGTFSWGNYKTLK-----EGVDDSSLHKELHKFYRDHYGSNRMVVALQAQLSLDELEEL 276
Query 225 IREKLEGISAGHADWLGMVQCPGPMFDTVKPFDHTNSGKFIHLQSFSSQPSLWVAFGLPA 284
+ I + + + Q + K F +Q L + + LP
Sbjct 277 LVRHCADIPTSQQNSIDVSQ-----LNYQKAFRDQFYKDVFLVQPVEDVCKLELTWVLPP 331
Query 285 TLTSYKKQPTSVLTYLLEYTGEGSLAKRLR----LLGLADGIS-PAVDRNSISTLLGIKV 339
Y+ +P ++ L+ Y G GSL LR + + G++ + D NSI +L I +
Sbjct 332 MKNFYRSKPDMFISQLIGYEGVGSLCAYLRHHLWCISVVAGVAESSFDSNSIYSLFNICI 391
Query 340 DLTQKGAAHRGLVLQEIFSYINFL 363
L+ G H VL+ F+++ +
Sbjct 392 YLSDDGFDHIDEVLEATFAWVKLI 415
> At3g02090
Length=531
Score = 73.2 bits (178), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 53/191 (27%), Positives = 88/191 (46%), Gaps = 10/191 (5%)
Query 21 RHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPEP 80
R L NG+ + + A V + GS ++ ++ G AHFLEHM+F GT +
Sbjct 98 RVTTLPNGLRVATESNLSAKTATVGVWIDAGSRFESDETNGTAHFLEHMIFKGTDRR-TV 156
Query 81 ESYDSFLTQNGGANNAYTDEEKTVFFNKVTDSAFEEALDRFAEFFKSPLFNRQYEEKEVN 140
+ + + GG NAYT E+T ++ KV DS +ALD A+ ++ F Q +E +
Sbjct 157 RALEEEIEDIGGHLNAYTSREQTTYYAKVLDSNVNQALDVLADILQNSKFEEQRINRERD 216
Query 141 AIDAEHQKNIPNDDERAWYTIRSLA--KGPMSRFATGNAETLSTYPKANGIDLVDRLRDF 198
I E Q+ DE + + A P+ R G A+ + + + + L+++
Sbjct 217 VILREMQEVEGQTDEVVLDHLHATAFQYTPLGRTILGPAQNVKSITRED-------LQNY 269
Query 199 HSKYYCGSNMV 209
+Y S MV
Sbjct 270 IKTHYTASRMV 280
> SPBP23A10.15c
Length=457
Score = 57.8 bits (138), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 49/176 (27%), Positives = 79/176 (44%), Gaps = 8/176 (4%)
Query 25 LSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPEPESYD 84
L NG+ HHP + A V + GS + G AHFLEH+ F GT +
Sbjct 27 LKNGLTVATEHHPYAQTATVLVGVDAGSRAETAKNNGAAHFLEHLAFKGTKNRSQKALEL 86
Query 85 SFLTQNGGAN-NAYTDEEKTVFFNKVTDSAFEEALDRFAEFFKSPLFNRQYEEKEVNAID 143
F +N GA+ NAYT E+TV++ +A A+ A+ + + E+E I
Sbjct 87 EF--ENTGAHLNAYTSREQTVYYAHAFKNAVPNAVAVLADILTNSSISASAVERERQVIL 144
Query 144 AEHQKNIPNDDERAWYTIRSLA-KG-PMSRFATGNAETLSTYPKANGIDLVDRLRD 197
E ++ DE + + + A +G P+ R G E + + + DL+ ++D
Sbjct 145 REQEEVDKMADEVVFDHLHATAYQGHPLGRTILGPKENIESLTRE---DLLQYIKD 197
> At5g42390
Length=1265
Score = 57.4 bits (137), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 62/229 (27%), Positives = 91/229 (39%), Gaps = 50/229 (21%)
Query 9 DILKPEADYRDFRHY--------------QLSNGMHAIAVHH---PRSNEAGFAVAANTG 51
D+L PE D + + QL NG+ + + + P EA V + G
Sbjct 170 DLLPPEIDSAELEAFLGCELPSHPKLHRGQLKNGLRYLILPNKVPPNRFEAHMEV--HVG 227
Query 52 SLYDPEDVPGLAHFLEHMLFLGTSKYPEPESYDSFLTQNGGANNAYTDEEKTVFF----- 106
S+ + ED G+AH +EH+ FLG+ K + L G +NAYTD TVF
Sbjct 228 SIDEEEDEQGIAHMIEHVAFLGSKKREK-------LLGTGARSNAYTDFHHTVFHIHSPT 280
Query 107 --NKVTDSAFEEALDRFAEFFKSPLFNRQYEEKEVNAIDAEHQK------NIPNDDERAW 158
D F LD E P F EKE AI +E Q + +
Sbjct 281 HTKDSEDDLFPSVLDALNEIAFHPKFLSSRVEKERRAILSELQMMNTIEYRVDCQLLQHL 340
Query 159 YTIRSLAKGPMSRFATGNAETLSTYPKANGIDLVDRLRDFHSKYYCGSN 207
++ L + RF G E + + VD++R FH ++Y +N
Sbjct 341 HSENKLGR----RFPIGLEEQIKKWD-------VDKIRKFHERWYFPAN 378
> YLR163c
Length=462
Score = 55.8 bits (133), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 45/191 (23%), Positives = 82/191 (42%), Gaps = 10/191 (5%)
Query 21 RHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPEP 80
R +L NG+ + P ++ A + + GS + G AHFLEH+ F GT +
Sbjct 27 RTSKLPNGLTIATEYIPNTSSATVGIFVDAGSRAENVKNNGTAHFLEHLAFKGTQNRSQ- 85
Query 81 ESYDSFLTQNGGANNAYTDEEKTVFFNKVTDSAFEEALDRFAEFFKSPLFNRQYEEKEVN 140
+ + + G NAYT E TV++ K +A+D ++ + + E+E +
Sbjct 86 QGIELEIENIGSHLNAYTSRENTVYYAKSLQEDIPKAVDILSDILTKSVLDNSAIERERD 145
Query 141 AIDAEHQKNIPNDDERAWYTIRSLA--KGPMSRFATGNAETLSTYPKANGIDLVDRLRDF 198
I E ++ DE + + + P+ R G + + + + + L+D+
Sbjct 146 VIIRESEEVDKMYDEVVFDHLHEITYKDQPLGRTILGPIKNIKSITRTD-------LKDY 198
Query 199 HSKYYCGSNMV 209
+K Y G MV
Sbjct 199 ITKNYKGDRMV 209
> Hs4758734
Length=489
Score = 54.7 bits (130), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 50/175 (28%), Positives = 77/175 (44%), Gaps = 12/175 (6%)
Query 49 NTGSLYDPEDVPGLAHFLEHMLFLGTSKYPEPESYDSFLTQNGGAN-NAYTDEEKTVFFN 107
+ GS Y+ E G AHFLEHM F GT K + + +N GA+ NAYT E+TV++
Sbjct 86 DAGSRYENEKNNGTAHFLEHMAFKGTKKRSQLDL--ELEIENMGAHLNAYTSREQTVYYA 143
Query 108 KVTDSAFEEALDRFAEFFKSPLFNRQYEEKEVNAIDAEHQKNIPNDDERAWYTIRSLA-- 165
K A++ A+ ++ E+E I E Q+ N E + + + A
Sbjct 144 KAFSKDLPRAVEILADIIQNSTLGEAEIERERGVILREMQEVETNLQEVVFDYLHATAYQ 203
Query 166 KGPMSRFATGNAETLSTYPKANGIDLVDRLRDFHSKYYCGSNMVAVTISPRSLDE 220
+ R G E + + + DLV D+ + +Y G +V S DE
Sbjct 204 NTALGRTILGPTENIKSISRK---DLV----DYITTHYKGPRIVLAAAGGVSHDE 251
> CE03806
Length=485
Score = 52.4 bits (124), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 39/118 (33%), Positives = 53/118 (44%), Gaps = 8/118 (6%)
Query 25 LSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPEPESYD 84
L NG +A + + A V + GS Y+ E G AHFLEHM F GT +
Sbjct 62 LPNGFR-VATENTGGSTATIGVFIDAGSRYENEKNNGTAHFLEHMAFKGTPR--RTRMGL 118
Query 85 SFLTQNGGAN-NAYTDEEKTVFFNKVTDSAFEEALDRFAEFFKSPLFNRQYEEKEVNA 141
+N GA+ NAYT E T ++ K F E LD+ + L N K++ A
Sbjct 119 ELEVENIGAHLNAYTSRESTTYYAK----CFTEKLDQSVDILSDILLNSSLATKDIEA 172
> At5g56730
Length=956
Score = 52.0 bits (123), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 48/203 (23%), Positives = 89/203 (43%), Gaps = 24/203 (11%)
Query 24 QLSNGMHAIAVHHPRSN-----EAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 78
+L NG+ +++ R N A A+A GS+ + ED G+AH +EH+ F T++Y
Sbjct 44 RLDNGL----IYYVRRNSKPRMRAALALAVKVGSVLEEEDQRGVAHIVEHLAFSATTRYT 99
Query 79 EPESY---DSFLTQNGGANNAYTDEEKTVF--FNKVTD-SAFEEALDRFAEFFKSPLFNR 132
+ +S + G NA T ++T++ F V +A+ AEF ++
Sbjct 100 NHDIVKFLESIGAEFGPCQNAMTTADETIYELFVPVDKPELLSQAISILAEFSSEIRVSK 159
Query 133 QYEEKEVNAIDAEHQ--KNIPNDDERAWYTIRSLAKGPMSRFATGNAETLSTYPKANGID 190
+ EKE A+ E++ +N + + + + R G + + + P A
Sbjct 160 EDLEKERGAVMEEYRGNRNATGRMQDSHWQLMMEGSKYAERLPIGLEKVIRSVPAAT--- 216
Query 191 LVDRLRDFHSKYYCGSNMVAVTI 213
++ F+ K+Y NM V +
Sbjct 217 ----VKQFYQKWYHLCNMAVVAV 235
> CE03926
Length=995
Score = 47.4 bits (111), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 41/169 (24%), Positives = 72/169 (42%), Gaps = 22/169 (13%)
Query 61 GLAHFLEHMLFLGTSKYPEPESYDSFLTQN-GGANNAYTDEEKTVF-FNKVTDSAFEEAL 118
GL H LEH++F+G+ KYP D + NA+TD + T + + V F + L
Sbjct 57 GLPHTLEHLVFMGSKKYPFKGVLDVIANRCLADGTNAWTDTDHTAYTLSTVGSDGFLKVL 116
Query 119 DRFAEFFKSPLFNRQYEEKEVNAIDAE-------------HQKNIPNDDERAWYTIRSLA 165
+ +P+ EV+ I E H+ + + +R + +
Sbjct 117 PVYINHLLTPMLTASQFATEVHHITGEGNDAGVVYSEMQDHESEMESIMDRK---TKEVI 173
Query 166 KGPMSRFATGNAETLSTYPKANGIDLVDRLRDFHSKYYCGSNMVAVTIS 214
P + +A L ++ + +++RD+H K+Y SNMV VT+
Sbjct 174 YPPFNPYAVDTGGRLKNLRESCTL---EKVRDYHKKFYHLSNMV-VTVC 218
> Hs4507841
Length=480
Score = 45.8 bits (107), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 47/216 (21%), Positives = 88/216 (40%), Gaps = 11/216 (5%)
Query 25 LSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPEPESYD 84
L NG+ +A V + GS ++ E G +FLEH+ F GT P + +
Sbjct 53 LDNGLR-VASEQSSQPTCTVGVWIDVGSRFETEKNNGAGYFLEHLAFKGTKNRPG-SALE 110
Query 85 SFLTQNGGANNAYTDEEKTVFFNKVTDSAFEEALDRFAEFFKSPLFNRQYEEKEVNAIDA 144
+ G NAY+ E T ++ K +A++ + ++ EKE + I
Sbjct 111 KEVESMGAHLNAYSTREHTAYYIKALSKDLPKAVELLGDIVQNCSLEDSQIEKERDVILR 170
Query 145 EHQKNIPNDDERAWYTIRSLA--KGPMSRFATGNAETLSTYPKANGIDLVDRLRDFHSKY 202
E Q+N + + + + + A P+++ G +E + +A+ L ++ S +
Sbjct 171 EMQENDASMRDVVFNYLHATAFQGTPLAQAVEGPSENVRKLSRAD-------LTEYLSTH 223
Query 203 YCGSNMVAVTISPRSLDEQESLIREKLEGISAGHAD 238
Y MV + L ++ L GI +A+
Sbjct 224 YKAPRMVLAAAGGVEHQQLLDLAQKHLGGIPWTYAE 259
> SPAC3H1.02c
Length=1036
Score = 43.5 bits (101), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 43/197 (21%), Positives = 78/197 (39%), Gaps = 18/197 (9%)
Query 26 SNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPEPESYDS 85
+ G +V P S G V A + D G H LEH+ F+G+ KYP
Sbjct 28 ATGFSVASVKTPTSRLQGSFVVAT-----EAHDNLGCPHTLEHLCFMGSKKYPMNGILTK 82
Query 86 FLTQNGGANNAYTDEEKTVF-FNKVTDSAFEEALDRFAEFFKSPLFNRQYEEKEVNAIDA 144
F + G NA TD + T + + + F L FA+ SP+ + + EV I+
Sbjct 83 FAGRACGDINACTDVDYTSYELSAAEEDGFLRLLPVFADHILSPILSDEAFCTEVYHING 142
Query 145 ---------EHQKNIPNDDERAWYTIRSLAKGPMSRFATGNAETLSTYPKANGIDLVDRL 195
+N + + + ++ P++ +G +P ++++
Sbjct 143 MGEESGVVYSEMQNTQSSETDVMFDCMRTSQYPVT---SGYYYETGGHPSELRKLSIEKI 199
Query 196 RDFHSKYYCGSNMVAVT 212
R++H + Y SN+ +
Sbjct 200 REYHKEMYVPSNICLIV 216
> YOL098c
Length=1037
Score = 42.4 bits (98), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 45/186 (24%), Positives = 72/186 (38%), Gaps = 30/186 (16%)
Query 61 GLAHFLEHMLFLGTSKYPEPESYDSFLTQNGGANNAYTDEEKTVF-FNKVTDSAFEEALD 119
G H LEH++F+G+ YP D+ + NA+TD ++TV+ + F + L
Sbjct 57 GAPHTLEHLIFMGSKSYPYKGLLDTAGNLSLSNTNAWTDTDQTVYTLSSAGWKGFSKLLP 116
Query 120 RFAEFFKSPLFNRQYEEKEVNAIDAEHQKN-------IPNDDERAWYTIRSLAK------ 166
+ + P + EV ID E+ + + + + WY I L K
Sbjct 117 AYLDHILHPTLTDEACLTEVYHIDPENLGDKGVVFSEMEAIETQGWY-ISGLEKQRLMFP 175
Query 167 -GPMSRFATG----NAETLSTYPKANGIDLVDRLRDFHSKYYCGSNMVAVTISPRSLDEQ 221
G R TG N TL+ D +R FH Y N+ + DE
Sbjct 176 EGSGYRSETGGLTKNLRTLTN----------DEIRQFHKSLYSSDNLCVIVCGNVPTDEL 225
Query 222 ESLIRE 227
+++ E
Sbjct 226 LTVMEE 231
> CE11226
Length=471
Score = 42.0 bits (97), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 33/121 (27%), Positives = 54/121 (44%), Gaps = 9/121 (7%)
Query 25 LSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPEPESYD 84
L NG + + S A V TGS ++ E G+AHFLE ++ GT K + +
Sbjct 43 LKNGFRVVTEDNG-SATATVGVWIETGSRFENEKNNGVAHFLERLIHKGTGKRASA-ALE 100
Query 85 SFLTQNGGANNAYTDEEKTVFFNKVTDSAFEEALDRFAEFFKSPLFNRQYEEKEVNAIDA 144
S L G N++T+ ++T F + E+ +D A+ ++ E + ID
Sbjct 101 SELNAIGAKLNSFTERDQTAVFVQAGAQDVEKVVDILADVLRNSKL-------EASTIDT 153
Query 145 E 145
E
Sbjct 154 E 154
> CE16673
Length=301
Score = 40.0 bits (92), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 21/73 (28%), Positives = 38/73 (52%), Gaps = 1/73 (1%)
Query 760 VQFQLGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEG 819
V +Q+G+ + D + ++ + +D+LRT+ LGYI + T LQ V+G
Sbjct 76 VTYQIGVQNTYDNAVIGLIKNLITEPAFDTLRTKESLGYIVWTRTHFNCGTVALQILVQG 135
Query 820 AKTHPDEVVKMID 832
K+ D V++ I+
Sbjct 136 PKS-VDHVLERIE 147
> YBR097w
Length=1454
Score = 40.0 bits (92), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 35/111 (31%), Positives = 52/111 (46%), Gaps = 16/111 (14%)
Query 47 AANTGSLYDPEDVPGLAHFLEHMLFLGTSK----YPEPESYDSFLTQNGGANNAYTDEEK 102
AA +Y PED PG E + + TSK Y PE ++S L Q+G +NN +E
Sbjct 167 AAFIKPVYLPEDNPG-----EFLFYFDTSKRRTCYLAPERFNSKLYQDGKSNNGRLTKEM 221
Query 103 TVF-FNKVTDSAFEEALDRF--AEFFK----SPLFNRQYEEKEVNAIDAEH 146
+F V F E F ++ FK S NR++ +E+N+ D +
Sbjct 222 DIFSLGCVIAEIFAEGRPIFNLSQLFKYKSNSYDVNREFLMEEMNSTDLRN 272
> Hs15321446
Length=525
Score = 39.7 bits (91), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 26/121 (21%), Positives = 51/121 (42%), Gaps = 1/121 (0%)
Query 25 LSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPEPESYD 84
L NG+ +A + + N+GS Y+ + + G+AHFLE + F T+++ +
Sbjct 72 LDNGLR-VASQNKFGQFCTVGILINSGSRYEAKYLSGIAHFLEKLAFSSTARFDSKDEIL 130
Query 85 SFLTQNGGANNAYTDEEKTVFFNKVTDSAFEEALDRFAEFFKSPLFNRQYEEKEVNAIDA 144
L ++GG + T + T++ + + A+ P + E A+
Sbjct 131 LTLEKHGGICDCQTSRDTTMYAVSADSKGLDTVVALLADVVLQPRLTDEEVEMTRMAVQF 190
Query 145 E 145
E
Sbjct 191 E 191
> At3g19170
Length=1052
Score = 36.6 bits (83), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 52/208 (25%), Positives = 87/208 (41%), Gaps = 33/208 (15%)
Query 20 FRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPE 79
F+H + G ++V + N+ F V T P+D G+ H LEH + G+ KYP
Sbjct 97 FKHKK--TGCEVMSVSNEDENKV-FGVVFRT----PPKDSTGIPHILEHSVLCGSRKYPV 149
Query 80 PESYDSFLTQNGGAN---NAYTDEEKTVFFNKVTDSA-FEEALDRF--AEFFKSPLFNRQ 133
E + L G + NA+T ++T + T++ F +D + A FF + +
Sbjct 150 KEPFVELL--KGSLHTFLNAFTYPDRTCYPVASTNTKDFYNLVDVYLDAVFFPKCVDDAH 207
Query 134 YEEKEVNAIDAEHQKNIPNDDERAWYTIRSLAKGPMS-------RFATGNAETLSTYPKA 186
++E ++ N P++D + + KG S R A +TY
Sbjct 208 TFQQE----GWHYELNDPSEDISYKGVVFNEMKGVYSQPDNILGRIAQQALSPENTYGVD 263
Query 187 NGIDLVD-------RLRDFHSKYYCGSN 207
+G D D ++FH +YY SN
Sbjct 264 SGGDPKDIPNLTFEEFKEFHRQYYHPSN 291
> 7304131
Length=556
Score = 36.2 bits (82), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 23/110 (20%), Positives = 46/110 (41%), Gaps = 3/110 (2%)
Query 25 LSNGMHAIAVHHPRSNE-AGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPEPESY 83
L NG+ + PR + + ++G Y+ G++HFLE + F T +P ++
Sbjct 99 LPNGLRIAS--EPRYGQFCTVGLVIDSGPRYEVAYPSGVSHFLEKLAFNSTVNFPNKDAI 156
Query 84 DSFLTQNGGANNAYTDEEKTVFFNKVTDSAFEEALDRFAEFFKSPLFNRQ 133
L +NGG + + + ++ + A + A+ P + Q
Sbjct 157 LKELEKNGGICDCQSSRDTLIYAASIDSRAIDSVTRLLADVTLRPTLSDQ 206
> At1g49630
Length=1076
Score = 35.4 bits (80), Expect = 0.53, Method: Compositional matrix adjust.
Identities = 25/89 (28%), Positives = 42/89 (47%), Gaps = 12/89 (13%)
Query 20 FRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPE 79
F+H + G ++V + N+ F + T P+D G+ H LEH + G+ KYP
Sbjct 124 FKHKK--TGCEVMSVSNDDENKV-FGIVFRT----PPKDSTGIPHILEHSVLCGSRKYPM 176
Query 80 PESYDSFLTQNGGAN---NAYTDEEKTVF 105
E + L G + NA+T ++T +
Sbjct 177 KEPFVELL--KGSLHTFLNAFTYPDRTCY 203
> Hs20535969
Length=417
Score = 35.4 bits (80), Expect = 0.54, Method: Compositional matrix adjust.
Identities = 23/98 (23%), Positives = 43/98 (43%), Gaps = 4/98 (4%)
Query 45 AVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPEPESYDSFLTQNGGANNAYTDEEKTV 104
+ N+GS Y+ + G+AHFLE + F +++ + L ++GG + T + T
Sbjct 39 GILINSGSRYEATYLSGIAHFLEKLAFSSNARFNSKDEILLTLEKHGGICDCQTSSDTTT 98
Query 105 FFNKVTDSAFEEALDRFAEFFKSPLFNRQYEEKEVNAI 142
+ SA + LD + Q ++EV +
Sbjct 99 Y----AVSADSKGLDTVFGLLADAVLQPQLTDEEVEMM 132
> At5g40740
Length=752
Score = 34.3 bits (77), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 63/224 (28%), Positives = 97/224 (43%), Gaps = 46/224 (20%)
Query 265 IHLQSFSSQPSLWVAFGLPATLT--SYKKQPTSVLTYLLEYTGEGSLAKRLRLLGLADGI 322
+H ++F P+ + LP++LT S+ T LL T + +R R L A+
Sbjct 175 VHRRTF---PADVASNPLPSSLTDVSFSHAAT-----LLPVTKARIVLERRRFLKNAE-- 224
Query 323 SPAVDRNSISTLLGIKVDLTQKGAAHRGLVLQEIFSYINFLRDH-----GVGHDLVSTMA 377
AV R ++ + L ++ +G LQ+ +N LR+ V DLVS+ +
Sbjct 225 -TAVQRQAMWSNLAHEMTAEFRGLCAEEAYLQQELEKLNDLRNKVKQEGEVWDDLVSSSS 283
Query 378 QQSHIDFHTTQPSSSIMDEAARLAHNLLTYEPY----------HVVAGDSLLIDAD---- 423
Q SH+ T+ SIM A + H +L P + ++G +LL D
Sbjct 284 QNSHLVSKATRLWDSIM--ARKGQHEVLASGPIEDLIAHREHRYRISGSALLAAMDQSSQ 341
Query 424 -PRLTNQLLQAMSPSKAIIA----FSDPDFT-----SKVDNFET 457
PR +LL A S A +A SD +T S VD+FET
Sbjct 342 VPRA--ELLSAHSDDSASLADDKELSDGSYTNMHDHSLVDSFET 383
> YDR430c
Length=989
Score = 33.9 bits (76), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 50/203 (24%), Positives = 81/203 (39%), Gaps = 22/203 (10%)
Query 44 FAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPEPESYDSFLTQN-GGANNAYTDEEK 102
F++A T +P D G+ H LEH G+ KYP + + L ++ NA T +
Sbjct 68 FSIAFKT----NPPDSTGVPHILEHTTLCGSVKYPVRDPFFKMLNKSLANFMNAMTGPDY 123
Query 103 TVF-FNKVTDSAFEEALDRFAEFFKSPLFNRQYEEKEVNAIDAEHQKNIPNDDERAWY-- 159
T F F+ F + + +PL ++ ++E EH KNI + + +
Sbjct 124 TFFPFSTTNPQDFANLRGVYLDSTLNPLLKQEDFDQE--GWRLEH-KNITDPESNIVFKG 180
Query 160 TIRSLAKGPMS--RFATGNAETLSTYPKANG-----IDLVD----RLRDFHSKYYCGSNM 208
+ + KG +S + + S YP N + + D L DFH K Y SN
Sbjct 181 VVYNEMKGQISNANYYFWSKFQQSIYPSLNNSGGDPMKITDLRYGDLLDFHHKNYHPSNA 240
Query 209 VAVTISPRSLDEQESLIREKLEG 231
T L + + E+ G
Sbjct 241 KTFTYGNLPLVDTLKQLNEQFSG 263
> 7299936
Length=382
Score = 33.5 bits (75), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 40/169 (23%), Positives = 70/169 (41%), Gaps = 12/169 (7%)
Query 69 MLFLGTSKYPEPESYDSFLTQNGGAN-NAYTDEEKTVFFNKVTDSAFEEALDRFAEFFKS 127
M F GT+K + + +N GA+ NAYT E+TVF+ K +A++ A+ ++
Sbjct 1 MAFKGTAKRSQTDL--ELEVENLGAHLNAYTSREQTVFYAKCLSKDVPKAVEILADIIQN 58
Query 128 PLFNRQYEEKEVNAIDAEHQKNIPNDDERAWYTIRSLA--KGPMSRFATGNAETLSTYPK 185
+E + I E Q+ N E + + + A P+ + G + + + K
Sbjct 59 SKLGEAEIARERSVILREMQEVESNLQEVVFDHLHATAYQGTPLGQTILGPTKNIQSIGK 118
Query 186 ANGIDLVDRLRDFHSKYYCGSNMVAVTISPRSLDEQESLIREKLEGISA 234
A+ L D+ +Y S +V D+ L L G+ A
Sbjct 119 AD-------LTDYIQTHYKASRIVLAAAGGVKHDDLVKLACSSLGGLEA 160
> Hs7657343
Length=1038
Score = 32.0 bits (71), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 21/77 (27%), Positives = 35/77 (45%), Gaps = 2/77 (2%)
Query 56 PEDVPGLAHFLEHMLFLGTSKYPEPESYDSFLTQNGGA-NNAYTDEEKTVF-FNKVTDSA 113
P D G+ H LEH + G+ KYP + L ++ NA+T + T++ F+
Sbjct 96 PMDSTGVPHILEHTVLCGSQKYPCRNPFFKMLNRSLSTFMNAFTASDYTLYPFSTQNPKD 155
Query 114 FEEALDRFAEFFKSPLF 130
F+ L + + SP
Sbjct 156 FQNLLSVYLDATFSPCL 172
> Hs17933772
Length=103
Score = 32.0 bits (71), Expect = 5.9, Method: Composition-based stats.
Identities = 18/66 (27%), Positives = 33/66 (50%), Gaps = 3/66 (4%)
Query 107 NKVTDSAFEEALDRFAEFFKSPLFNRQYEEKEVNAIDAEHQKNIPNDDERAWYTIRSLAK 166
NK++ S+F E L + S NR+ +K + +DA H I D+ ++T+
Sbjct 31 NKISKSSFREMLQKELNHMLSDTGNRKAADKLIQNLDANHDGRISFDE---YWTLIGGIT 87
Query 167 GPMSRF 172
GP+++
Sbjct 88 GPIAKL 93
> 7302255
Length=1112
Score = 31.6 bits (70), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 26/51 (50%), Gaps = 1/51 (1%)
Query 56 PEDVPGLAHFLEHMLFLGTSKYPEPESYDSFLTQNGGA-NNAYTDEEKTVF 105
P D GL H LEH+ G+ KYP + + L ++ NA T + T++
Sbjct 120 PFDSTGLPHILEHLSLCGSQKYPVRDPFFKMLNRSVATFMNAMTGPDYTIY 170
Lambda K H
0.317 0.132 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 25305168348
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40