bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
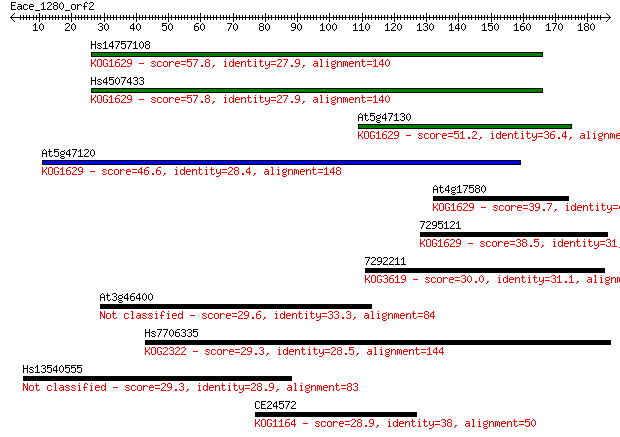
Query= Eace_1280_orf2
Length=186
Score E
Sequences producing significant alignments: (Bits) Value
Hs14757108 57.8 1e-08
Hs4507433 57.8 1e-08
At5g47130 51.2 1e-06
At5g47120 46.6 3e-05
At4g17580 39.7 0.004
7295121 38.5 0.008
7292211 30.0 3.1
At3g46400 29.6 3.8
Hs7706335 29.3 4.4
Hs13540555 29.3 4.7
CE24572 28.9 5.9
> Hs14757108
Length=237
Score = 57.8 bits (138), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 39/140 (27%), Positives = 72/140 (51%), Gaps = 5/140 (3%)
Query 26 SLLDFTPLSAPQKRHVSKIYAALTVNVLLTAVGVYAQLRWVSLPTFLSLLLSIGCVFGLT 85
+LL F+ ++ ++H+ K+YA+ + + + A G Y + V+ LL ++G + +
Sbjct 12 ALLKFSHITPSTQQHLKKVYASFALCMFVAAAGAYVHM--VTHFIQAGLLSALGSLILMI 69
Query 86 SSSQKAHAESQMLTKERALCSGGFGVLNGMLIANYLHAIHFYVGPQVIPTAFFASVAVFF 145
H+ ++R GF L G+ + L V P ++PTAF + +F
Sbjct 70 WLMATPHSHET--EQKRLGLLAGFAFLTGVGLGPALEFC-IAVNPSILPTAFMGTAMIFT 126
Query 146 CFSAAALVAKQRSYLYLGSL 165
CF+ +AL A++RSYL+LG +
Sbjct 127 CFTLSALYARRRSYLFLGGI 146
> Hs4507433
Length=237
Score = 57.8 bits (138), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 39/140 (27%), Positives = 72/140 (51%), Gaps = 5/140 (3%)
Query 26 SLLDFTPLSAPQKRHVSKIYAALTVNVLLTAVGVYAQLRWVSLPTFLSLLLSIGCVFGLT 85
+LL F+ ++ ++H+ K+YA+ + + + A G Y + V+ LL ++G + +
Sbjct 12 ALLKFSHITPSTQQHLKKVYASFALCMFVAAAGAYVHM--VTHFIQAGLLSALGSLILMI 69
Query 86 SSSQKAHAESQMLTKERALCSGGFGVLNGMLIANYLHAIHFYVGPQVIPTAFFASVAVFF 145
H+ ++R GF L G+ + L V P ++PTAF + +F
Sbjct 70 WLMATPHSHET--EQKRLGLLAGFAFLTGVGLGPALEFC-IAVNPSILPTAFMGTAMIFT 126
Query 146 CFSAAALVAKQRSYLYLGSL 165
CF+ +AL A++RSYL+LG +
Sbjct 127 CFTLSALYARRRSYLFLGGI 146
> At5g47130
Length=187
Score = 51.2 bits (121), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 24/66 (36%), Positives = 41/66 (62%), Gaps = 1/66 (1%)
Query 109 FGVLNGMLIANYLHAIHFYVGPQVIPTAFFASVAVFFCFSAAALVAKQRSYLYLGSLLGA 168
FGVL+G + + + + ++ TAF + +FFCFSA A++A++R Y+YLG LL +
Sbjct 40 FGVLHGASVGPCIKST-IDIDSSILITAFLGTAVIFFCFSAVAMLARRREYIYLGGLLSS 98
Query 169 ALTYLS 174
+ L+
Sbjct 99 GFSLLT 104
> At5g47120
Length=247
Score = 46.6 bits (109), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 42/154 (27%), Positives = 71/154 (46%), Gaps = 16/154 (10%)
Query 11 MEAFNFTGFSAVPA------RSLLDFTPLSAPQKRHVSKIYAALTVNVLLTAVGVYAQLR 64
M+AF+ + F + P SL +F +S + H+ ++Y L ++ +A G Y +
Sbjct 1 MDAFS-SFFDSQPGSRSWSYDSLKNFRQISPAVQNHLKRVYLTLCCALVASAFGAYLHVL 59
Query 65 WVSLPTFLSLLLSIGCVFGLTSSSQKAHAESQMLTKERALCSGGFGVLNGMLIANYLHAI 124
W ++ L+ + IG + L S H + L A VL G + L +
Sbjct 60 W-NIGGILTTIGCIGTMIWLLSCPPYEHQKRLSLLFVSA-------VLEGASVGP-LIKV 110
Query 125 HFYVGPQVIPTAFFASVAVFFCFSAAALVAKQRS 158
V P ++ TAF + F CFSAAA++A++R
Sbjct 111 AIDVDPSILITAFVGTAIAFVCFSAAAMLARRRE 144
> At4g17580
Length=247
Score = 39.7 bits (91), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 19/42 (45%), Positives = 27/42 (64%), Gaps = 0/42 (0%)
Query 132 VIPTAFFASVAVFFCFSAAALVAKQRSYLYLGSLLGAALTYL 173
V+ TAF + F CFSAAA++A +R YLY G+ L ++ L
Sbjct 120 VLVTAFVGTAVAFVCFSAAAMLATRREYLYHGASLACCMSIL 161
> 7295121
Length=245
Score = 38.5 bits (88), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 18/58 (31%), Positives = 34/58 (58%), Gaps = 0/58 (0%)
Query 128 VGPQVIPTAFFASVAVFFCFSAAALVAKQRSYLYLGSLLGAALTYLSIASLVNIFLRA 185
+ P +I +A + F S +AL+A+Q YLYLG +L + + +++ SL N+ ++
Sbjct 112 INPAIILSALTGTFVTFISLSLSALLAEQGKYLYLGGMLVSVINTMALLSLFNMVFKS 169
> 7292211
Length=1180
Score = 30.0 bits (66), Expect = 3.1, Method: Composition-based stats.
Identities = 23/77 (29%), Positives = 40/77 (51%), Gaps = 6/77 (7%)
Query 111 VLNGMLIANYLHAIHFYVGPQVIPTAFFASVAVFFCF---SAAALVAKQRSYLYLGSLLG 167
+LN + A L+AI+ ++ + F AV FC+ + + AK + ++ S+L
Sbjct 155 ILNPIYDAYTLYAIYMFMPVPYLLQPFVLGSAVTFCYIINYSFVITAKDDNQMH--SILN 212
Query 168 AALTYLSIASLVNIFLR 184
A+ YLS +L+ IF R
Sbjct 213 EAI-YLSCVNLLGIFFR 228
> At3g46400
Length=867
Score = 29.6 bits (65), Expect = 3.8, Method: Composition-based stats.
Identities = 28/97 (28%), Positives = 45/97 (46%), Gaps = 13/97 (13%)
Query 29 DFTPLSAPQKRHVSKIYAALTVNVLLTAV------GVYAQLRW-----VSLPTFLSLLLS 77
D T LS K + AAL + ++ A+ V+ + +W V LPT + +
Sbjct 478 DNTCLSCVPKNKFPMMIAALAASAIVVAILVLILIFVFTKKKWSTHMEVILPTMDIMSKT 537
Query 78 IGCVFGLTSSSQKAHAESQMLTK--ERALCSGGFGVL 112
I T + A++E +TK E+AL GGFG++
Sbjct 538 ISEQLIKTKRRRFAYSEVVEMTKKFEKALGEGGFGIV 574
> Hs7706335
Length=258
Score = 29.3 bits (64), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 41/153 (26%), Positives = 70/153 (45%), Gaps = 22/153 (14%)
Query 43 KIYAALTVNVLLTAVGVYAQLRWVSLPTFL----SLLL-----SIGCVFGLTSSSQKAHA 93
K+Y+ L++ VLLT V L + S+ TF+ +L+L S+G +F LT + K
Sbjct 57 KVYSILSLQVLLTTVTSTVFLYFESVRTFVHESPALILLFALGSLGLIFALTLNRHKYPL 116
Query 94 ESQMLTKERALCSGGFGVLNGMLIANYLHAIHFYVGPQVIPTAFFASVAVFFCFSAAALV 153
+L GF +L + +A + FY +I AF + VFF + L
Sbjct 117 NLYLLF--------GFTLLEALTVAV---VVTFY-DVYIILQAFILTTTVFFGLTVYTLQ 164
Query 154 AKQRSYLYLGSLLGAALTYLSIASLVNIFLRAQ 186
+K + + G+ L A L L ++ + F ++
Sbjct 165 SK-KDFSKFGAGLFALLWILCLSGFLKFFFYSE 196
> Hs13540555
Length=487
Score = 29.3 bits (64), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 24/85 (28%), Positives = 38/85 (44%), Gaps = 3/85 (3%)
Query 5 HFVLLKMEAFNFTGFSAVPARSLLDFTPLSAPQKRHVSKIY--AALTVNVLLTAVGVYAQ 62
H+++L F F V R + DF P K+ + + AA+T LL V
Sbjct 344 HYLVLLRLVF-FVNVGGVAMREIYDFMDDPKPHKKLGPQAWLVAAITATELLIVVKYDPH 402
Query 63 LRWVSLPTFLSLLLSIGCVFGLTSS 87
+SLP ++S ++G V LT +
Sbjct 403 TLTLSLPFYISQCWTLGSVLALTWT 427
> CE24572
Length=382
Score = 28.9 bits (63), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 19/52 (36%), Positives = 27/52 (51%), Gaps = 5/52 (9%)
Query 77 SIGCVFGLTSSSQKAH--AESQMLTKERALCSGGFGVLNGMLIANYLHAIHF 126
S G VFG+T S K H + + + +RA+ GVL L N+L + HF
Sbjct 80 SFGAVFGVTRKSDKRHFAMKCESVNMKRAILPHEAGVL---LALNHLKSPHF 128
Lambda K H
0.328 0.138 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3022542264
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40