bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1310_orf1
Length=108
Score E
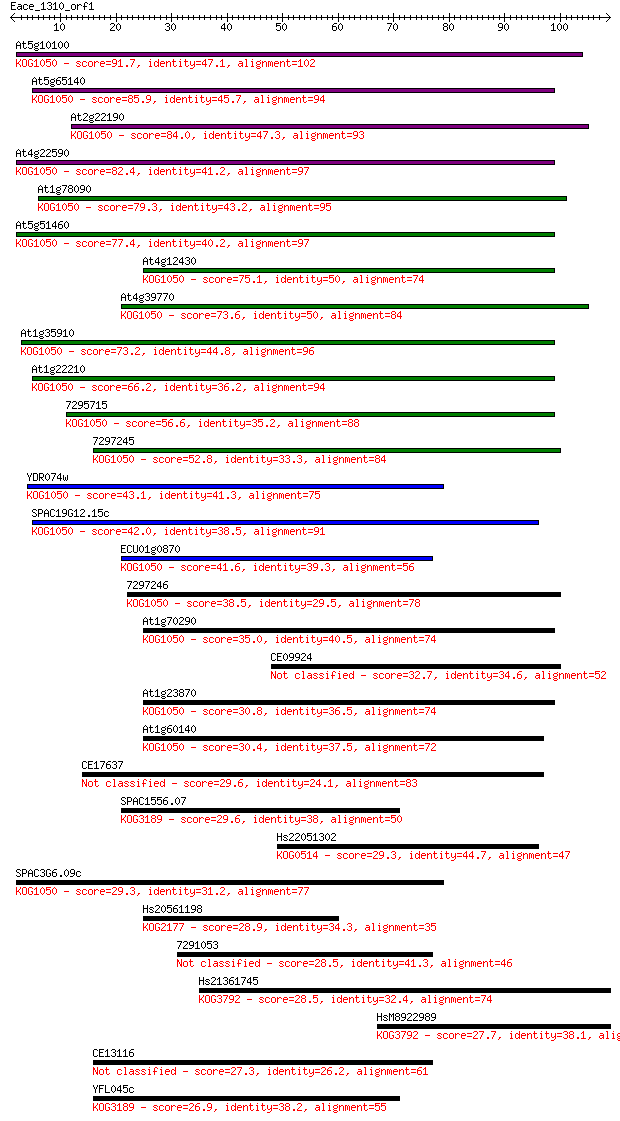
Sequences producing significant alignments: (Bits) Value
At5g10100 91.7 3e-19
At5g65140 85.9 2e-17
At2g22190 84.0 6e-17
At4g22590 82.4 2e-16
At1g78090 79.3 1e-15
At5g51460 77.4 6e-15
At4g12430 75.1 3e-14
At4g39770 73.6 8e-14
At1g35910 73.2 1e-13
At1g22210 66.2 1e-11
7295715 56.6 9e-09
7297245 52.8 2e-07
YDR074w 43.1 1e-04
SPAC19G12.15c 42.0 3e-04
ECU01g0870 41.6 3e-04
7297246 38.5 0.003
At1g70290 35.0 0.033
CE09924 32.7 0.17
At1g23870 30.8 0.61
At1g60140 30.4 0.82
CE17637 29.6 1.3
SPAC1556.07 29.6 1.5
Hs22051302 29.3 1.6
SPAC3G6.09c 29.3 1.9
Hs20561198 28.9 2.6
7291053 28.5 3.0
Hs21361745 28.5 3.3
HsM8922989 27.7 5.9
CE13116 27.3 7.4
YFL045c 26.9 9.0
> At5g10100
Length=372
Score = 91.7 bits (226), Expect = 3e-19, Method: Composition-based stats.
Identities = 48/104 (46%), Positives = 67/104 (64%), Gaps = 8/104 (7%)
Query 2 LETPNLLDEVQYVRYKLADKSLCIFLDYDGTLTPIVADPREAKMSPAFRKTLKKLARVFP 61
++ P+ L++ + + K + +FLDYDGTL+PIV DP +A MS R+T+KKLA+ FP
Sbjct 92 MQHPSALEKFEQIMEASRGKQIVMFLDYDGTLSPIVDDPDKAFMSSKMRRTVKKLAKCFP 151
Query 62 VAIVTGRRLETIRSFVGLADAPGRRALMYAASHGFDID--AAGF 103
AIVTGR ++ + +FV LA+ L YA SHG DI A GF
Sbjct 152 TAIVTGRCIDKVYNFVKLAE------LYYAGSHGMDIKGPAKGF 189
> At5g65140
Length=381
Score = 85.9 bits (211), Expect = 2e-17, Method: Composition-based stats.
Identities = 43/94 (45%), Positives = 61/94 (64%), Gaps = 6/94 (6%)
Query 5 PNLLDEVQYVRYKLADKSLCIFLDYDGTLTPIVADPREAKMSPAFRKTLKKLARVFPVAI 64
P+ L+ + + + K + +FLDYDGTL+PIV DP A M+ R+T+KK+A+ FP +I
Sbjct 98 PSALNMFERIIEEARGKQIVMFLDYDGTLSPIVDDPDRAFMTSKMRRTVKKMAKCFPTSI 157
Query 65 VTGRRLETIRSFVGLADAPGRRALMYAASHGFDI 98
VTGR ++ + SFV LA+ L YA SHG DI
Sbjct 158 VTGRCIDKVYSFVKLAE------LYYAGSHGMDI 185
> At2g22190
Length=269
Score = 84.0 bits (206), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 44/93 (47%), Positives = 56/93 (60%), Gaps = 6/93 (6%)
Query 12 QYVRYKLADKSLCIFLDYDGTLTPIVADPREAKMSPAFRKTLKKLARVFPVAIVTGRRLE 71
+ + +K K + +FLDYDGTL+PIV DP A MS R T++KLA+ FP AIV+GR E
Sbjct 3 EEILHKSEGKQIVMFLDYDGTLSPIVDDPDRAFMSKKMRNTVRKLAKCFPTAIVSGRCRE 62
Query 72 TIRSFVGLADAPGRRALMYAASHGFDIDAAGFG 104
+ SFV L + L YA SHG DI G
Sbjct 63 KVSSFVKLTE------LYYAGSHGMDIKGPEQG 89
> At4g22590
Length=377
Score = 82.4 bits (202), Expect = 2e-16, Method: Composition-based stats.
Identities = 40/97 (41%), Positives = 58/97 (59%), Gaps = 6/97 (6%)
Query 2 LETPNLLDEVQYVRYKLADKSLCIFLDYDGTLTPIVADPREAKMSPAFRKTLKKLARVFP 61
L+ P+ + ++ + +K + +FLDYDGTL+PIV DP A MS A R +K +A+ FP
Sbjct 87 LKYPSAITSFAHIAAQAKNKKIAVFLDYDGTLSPIVDDPDRAIMSDAMRAAVKDVAKYFP 146
Query 62 VAIVTGRRLETIRSFVGLADAPGRRALMYAASHGFDI 98
AI++GR + + VGL + L YA SHG DI
Sbjct 147 TAIISGRSRDKVYQLVGLTE------LYYAGSHGMDI 177
> At1g78090
Length=374
Score = 79.3 bits (194), Expect = 1e-15, Method: Composition-based stats.
Identities = 41/95 (43%), Positives = 58/95 (61%), Gaps = 9/95 (9%)
Query 6 NLLDEVQYVRYKLADKSLCIFLDYDGTLTPIVADPREAKMSPAFRKTLKKLARVFPVAIV 65
N+ DE+ K + +FLDYDGTL+PIV DP +A ++ R+ +K +A FP AIV
Sbjct 104 NMFDEIVNAA---KGKQIVMFLDYDGTLSPIVEDPDKAFITHEMREVVKDVASNFPTAIV 160
Query 66 TGRRLETIRSFVGLADAPGRRALMYAASHGFDIDA 100
TGR +E +RSFV + + + YA SHG DI+
Sbjct 161 TGRSIEKVRSFVQVNE------IYYAGSHGMDIEG 189
> At5g51460
Length=385
Score = 77.4 bits (189), Expect = 6e-15, Method: Composition-based stats.
Identities = 39/97 (40%), Positives = 57/97 (58%), Gaps = 6/97 (6%)
Query 2 LETPNLLDEVQYVRYKLADKSLCIFLDYDGTLTPIVADPREAKMSPAFRKTLKKLARVFP 61
L+ P+ L + + K + +FLDYDGTL+PIV +P A MS A R ++ +A+ FP
Sbjct 100 LKYPSALTSFEKIMSFAKGKRIALFLDYDGTLSPIVEEPDCAYMSSAMRSAVQNVAKYFP 159
Query 62 VAIVTGRRLETIRSFVGLADAPGRRALMYAASHGFDI 98
AI++GR + + FV L++ L YA SHG DI
Sbjct 160 TAIISGRSRDKVYEFVNLSE------LYYAGSHGMDI 190
> At4g12430
Length=368
Score = 75.1 bits (183), Expect = 3e-14, Method: Composition-based stats.
Identities = 37/74 (50%), Positives = 46/74 (62%), Gaps = 6/74 (8%)
Query 25 IFLDYDGTLTPIVADPREAKMSPAFRKTLKKLARVFPVAIVTGRRLETIRSFVGLADAPG 84
+FLDYDGTL+PIV DP A MS A R +K +A FP AI++GR + + VGL +
Sbjct 107 VFLDYDGTLSPIVDDPDRAIMSDAMRSAVKDVASYFPTAIISGRSRDKVYQLVGLTE--- 163
Query 85 RRALMYAASHGFDI 98
L YA SHG DI
Sbjct 164 ---LYYAGSHGMDI 174
> At4g39770
Length=267
Score = 73.6 bits (179), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 42/94 (44%), Positives = 53/94 (56%), Gaps = 16/94 (17%)
Query 21 KSLCIFLDYDGTLTPIVADPREAKMSPAFRKTLKKLARVFPVAIVTGRRLE--------- 71
K + +FLDYDGTL+PIV DP A MS R+T++KLA FP AIV+GR +E
Sbjct 12 KQIVMFLDYDGTLSPIVDDPDRAFMSRKMRRTVRKLANCFPTAIVSGRCIEKLIMIVLTT 71
Query 72 -TIRSFVGLADAPGRRALMYAASHGFDIDAAGFG 104
+ +FV L + L YA SHG DI G
Sbjct 72 FQVYNFVKLTE------LYYAGSHGMDIKGPEQG 99
> At1g35910
Length=366
Score = 73.2 bits (178), Expect = 1e-13, Method: Composition-based stats.
Identities = 43/96 (44%), Positives = 56/96 (58%), Gaps = 9/96 (9%)
Query 3 ETPNLLDEVQYVRYKLADKSLCIFLDYDGTLTPIVADPREAKMSPAFRKTLKKLARVFPV 62
+ P+ L + + K + +FLDYDGTL+PIV +P A MS + +K +AR FP
Sbjct 93 QHPSALTMFEEIAEASKGKQIVMFLDYDGTLSPIVENPDRAYMS---EEAVKGVARYFPT 149
Query 63 AIVTGRRLETIRSFVGLADAPGRRALMYAASHGFDI 98
AIVTGR + +R FV L PG L YA SHG DI
Sbjct 150 AIVTGRCRDKVRRFVKL---PG---LYYAGSHGMDI 179
> At1g22210
Length=320
Score = 66.2 bits (160), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 34/94 (36%), Positives = 52/94 (55%), Gaps = 6/94 (6%)
Query 5 PNLLDEVQYVRYKLADKSLCIFLDYDGTLTPIVADPREAKMSPAFRKTLKKLARVFPVAI 64
P+ LD + + K + +FLDYDGTL+ I D A ++ R+ +K++A F AI
Sbjct 49 PSALDMFEQIMRDAEGKQIIMFLDYDGTLSLITEDHDRAYITDEMREVVKEVATYFKTAI 108
Query 65 VTGRRLETIRSFVGLADAPGRRALMYAASHGFDI 98
++GR + ++SFV L + YA SHG DI
Sbjct 109 ISGRSTDKVQSFVKLT------GIHYAGSHGMDI 136
> 7295715
Length=809
Score = 56.6 bits (135), Expect = 9e-09, Method: Composition-based stats.
Identities = 31/91 (34%), Positives = 51/91 (56%), Gaps = 13/91 (14%)
Query 11 VQYVRYKLADKSLCIFLDYDGTLTPIVADPREAKMSPAFRKTLKKLAR---VFPVAIVTG 67
++Y+ Y + L + LDYDGTL PI P A +SP + L KL+ V+ VA+++G
Sbjct 522 LKYIGY---NHKLALLLDYDGTLAPIAPHPDLATLSPEIKNVLYKLSNHSDVY-VAVISG 577
Query 68 RRLETIRSFVGLADAPGRRALMYAASHGFDI 98
R ++ ++ VG+ + YA +HG +I
Sbjct 578 RNVDNVKKMVGI------EGITYAGNHGLEI 602
> 7297245
Length=273
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 28/87 (32%), Positives = 49/87 (56%), Gaps = 10/87 (11%)
Query 16 YKLADKSLCIFLDYDGTLTPIVADPREAKMSPAFRKTLKKLA---RVFPVAIVTGRRLET 72
Y +++ + + LDYDGTL PI +P + KM L K+A +VF +A+++GR L+
Sbjct 23 YLISESPVAVLLDYDGTLAPIADNPAKTKMPVELEAILHKIAKHPKVF-LAVISGRGLKD 81
Query 73 IRSFVGLADAPGRRALMYAASHGFDID 99
++ V + + YA +HG +I+
Sbjct 82 VQKQVNI------DGITYAGNHGLEIE 102
> YDR074w
Length=896
Score = 43.1 bits (100), Expect = 1e-04, Method: Composition-based stats.
Identities = 31/78 (39%), Positives = 41/78 (52%), Gaps = 5/78 (6%)
Query 4 TPNLLDEVQYVRYKLADKSLCIFLDYDGTLTPIVADPREAKMSPAFRKTLKKLARVFP-- 61
TP L V YK A + L +F DYDGTLTPIV DP A S L+KL P
Sbjct 554 TPALNRPVLLENYKQAKRRLFLF-DYDGTLTPIVKDPAAAIPSARLYTILQKLC-ADPHN 611
Query 62 -VAIVTGRRLETIRSFVG 78
+ I++GR + + ++G
Sbjct 612 QIWIISGRDQKFLNKWLG 629
> SPAC19G12.15c
Length=817
Score = 42.0 bits (97), Expect = 3e-04, Method: Composition-based stats.
Identities = 35/93 (37%), Positives = 44/93 (47%), Gaps = 8/93 (8%)
Query 5 PNLLDEVQYVRYKLADKSLCIFLDYDGTLTPIVADPREAKMSPAFRKTLKKLAR--VFPV 62
P L E Y A K L +DYDGTLTPIV DP A S L LA V
Sbjct 548 PTLTPEHALSVYSKASKRL-FMMDYDGTLTPIVRDPNAAVPSKKLLDNLATLAADPKNQV 606
Query 63 AIVTGRRLETIRSFVGLADAPGRRALMYAASHG 95
I++GR + +R++ + D G L +A HG
Sbjct 607 WIISGRDQQFLRNW--MDDIKG---LGLSAEHG 634
> ECU01g0870
Length=718
Score = 41.6 bits (96), Expect = 3e-04, Method: Composition-based stats.
Identities = 22/56 (39%), Positives = 31/56 (55%), Gaps = 0/56 (0%)
Query 21 KSLCIFLDYDGTLTPIVADPREAKMSPAFRKTLKKLARVFPVAIVTGRRLETIRSF 76
K+ + +DYDGTLT IVA P A + + L +L ++ V I TGR +E F
Sbjct 501 KARTLVMDYDGTLTNIVARPPMAAPTQEIKDLLIRLGKICRVVISTGRSVEDCDKF 556
> 7297246
Length=276
Score = 38.5 bits (88), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 23/85 (27%), Positives = 46/85 (54%), Gaps = 20/85 (23%)
Query 22 SLCIFLDYDGTLTPIVAD----PREAKMSPAFRKTLKKLA---RVFPVAIVTGRRLETIR 74
+ + LDYDGTL P+ + P++ +++ +KKLA ++F V + +GR L I+
Sbjct 30 QVALLLDYDGTLAPLTEELSVMPKDTEIN------IKKLAANEKIFMV-VFSGRELSEIK 82
Query 75 SFVGLADAPGRRALMYAASHGFDID 99
+ + + + YA +HG +++
Sbjct 83 NHLKFPN------VTYAGNHGLEVE 101
> At1g70290
Length=826
Score = 35.0 bits (79), Expect = 0.033, Method: Composition-based stats.
Identities = 30/79 (37%), Positives = 39/79 (49%), Gaps = 15/79 (18%)
Query 25 IFLDYDGTLTP---IVADPREAKMSPAFRKTLKKLAR--VFPVAIVTGRRLETIRSFVGL 79
IFLDYDGTL P IV DP +S LK L V IV+GR E++ +++
Sbjct 559 IFLDYDGTLVPESSIVQDPSNEVVS-----VLKALCEDPNNTVFIVSGRGRESLSNWLSP 613
Query 80 ADAPGRRALMYAASHGFDI 98
+ G AA HG+ I
Sbjct 614 CENLG-----IAAEHGYFI 627
> CE09924
Length=288
Score = 32.7 bits (73), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 18/53 (33%), Positives = 29/53 (54%), Gaps = 6/53 (11%)
Query 48 AFRKTLKKLARVFPVAIVTGRRLETIRSFVGLADAPGRRALM-YAASHGFDID 99
A+++T+KK FP+++VTG TI S D P + ++ YA G + D
Sbjct 93 AYKQTIKKTGEYFPLSLVTGISYYTITS-----DNPNEKVVLSYAMRVGVNED 140
> At1g23870
Length=867
Score = 30.8 bits (68), Expect = 0.61, Method: Composition-based stats.
Identities = 27/79 (34%), Positives = 37/79 (46%), Gaps = 15/79 (18%)
Query 25 IFLDYDGTLTP---IVADPREAKMSPAFRKTLKKLA--RVFPVAIVTGRRLETIRSFVGL 79
IFLDYDGTL P I+ P +S LK L V +V+GR E++ ++
Sbjct 594 IFLDYDGTLVPESSIIKTPNAEVLS-----VLKSLCGDPKNTVFVVSGRGWESLSDWLSP 648
Query 80 ADAPGRRALMYAASHGFDI 98
+ G AA HG+ I
Sbjct 649 CENLG-----IAAEHGYFI 662
> At1g60140
Length=861
Score = 30.4 bits (67), Expect = 0.82, Method: Composition-based stats.
Identities = 27/77 (35%), Positives = 36/77 (46%), Gaps = 15/77 (19%)
Query 25 IFLDYDGTLTP---IVADPREAKMSPAFRKTLKKLAR--VFPVAIVTGRRLETIRSFVGL 79
IFLDYDGTL P IV DP +S LK L + IV+GR ++ ++
Sbjct 594 IFLDYDGTLVPETSIVKDPSAEVIS-----ALKALCSDPNNTIFIVSGRGKVSLSEWLAP 648
Query 80 ADAPGRRALMYAASHGF 96
+ G AA HG+
Sbjct 649 CENLG-----IAAEHGY 660
> CE17637
Length=383
Score = 29.6 bits (65), Expect = 1.3, Method: Composition-based stats.
Identities = 20/83 (24%), Positives = 37/83 (44%), Gaps = 1/83 (1%)
Query 14 VRYKLADKSLCIFLDYDGTLTPIVADPREAKMSPAFRKTLKKLARVFPVAIVTGRRLETI 73
++ L + + + L+P K P + KTL+ + FP + G +L I
Sbjct 25 IKITLGSSHILEYFQKNSNLSPKCEINNSQKSQPNYEKTLQLMLFAFPSGGL-GNKLFEI 83
Query 74 RSFVGLADAPGRRALMYAASHGF 96
S G+A + R+A++ A + F
Sbjct 84 ISLHGIATSLQRKAVINATNPSF 106
> SPAC1556.07
Length=257
Score = 29.6 bits (65), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 19/50 (38%), Positives = 26/50 (52%), Gaps = 6/50 (12%)
Query 21 KSLCIFLDYDGTLTPIVADPREAKMSPAFRKTLKKLARVFPVAIVTGRRL 70
K+L +F D DGTLTP +SP +TL+ L +V + V G L
Sbjct 13 KTLVLF-DVDGTLTPA-----RLSVSPEMLETLQNLRKVVAIGFVGGSDL 56
> Hs22051302
Length=389
Score = 29.3 bits (64), Expect = 1.6, Method: Composition-based stats.
Identities = 21/51 (41%), Positives = 30/51 (58%), Gaps = 4/51 (7%)
Query 49 FRKTLKKLARVFPVA-IVTG-RRL--ETIRSFVGLADAPGRRALMYAASHG 95
FR + ++ ++ PVA ++ G RRL E + V LAD G AL Y+ SHG
Sbjct 152 FRVSSQRRSQAEPVARMLEGVRRLGPELLAHVVNLADGNGNTALHYSVSHG 202
> SPAC3G6.09c
Length=849
Score = 29.3 bits (64), Expect = 1.9, Method: Composition-based stats.
Identities = 24/79 (30%), Positives = 39/79 (49%), Gaps = 3/79 (3%)
Query 2 LETPNLLDEVQYVRYKLADKSLCIFLDYDGTLTPIVADPREAKMSPAFRKTLKKLARVFP 61
+ TP L + Y+ A + L LDYDGTL + +A + +TLK+LA
Sbjct 556 MTTPLLTYNILIKPYRNAKRRL-FLLDYDGTLIESARNSIDAVPTDRLLRTLKRLASDSR 614
Query 62 --VAIVTGRRLETIRSFVG 78
V I++GR + + ++G
Sbjct 615 NIVWILSGRSQKFMEEWMG 633
> Hs20561198
Length=684
Score = 28.9 bits (63), Expect = 2.6, Method: Composition-based stats.
Identities = 12/35 (34%), Positives = 19/35 (54%), Gaps = 0/35 (0%)
Query 25 IFLDYDGTLTPIVADPREAKMSPAFRKTLKKLARV 59
+FLDYD T+ +E KM P + +K A++
Sbjct 277 VFLDYDNTVASFYDASKEKKMEPKPQVVVKSTAKL 311
> 7291053
Length=1008
Score = 28.5 bits (62), Expect = 3.0, Method: Composition-based stats.
Identities = 19/59 (32%), Positives = 27/59 (45%), Gaps = 13/59 (22%)
Query 31 GTLTPIVA-------DPREAKMSPA------FRKTLKKLARVFPVAIVTGRRLETIRSF 76
G TP+V P EA M A RK+L L + P+++VT +L + R F
Sbjct 153 GVATPLVMASPVRRRSPIEATMDKAPGQDSNHRKSLNLLDKALPISVVTAPQLRSRRQF 211
> Hs21361745
Length=672
Score = 28.5 bits (62), Expect = 3.3, Method: Composition-based stats.
Identities = 24/76 (31%), Positives = 34/76 (44%), Gaps = 3/76 (3%)
Query 35 PIVADPREAKMSPAFRKTLKKLARVFPVAIVTGRRLETIR--SFVGLADAPGRRALMYAA 92
P A+ ++ K+ P K + A V V GR T+ +FVG APG A Y
Sbjct 578 PNAANNKKKKIIPQ-AKGVVNTAVSAAVQAVRGRGRGTLTRGAFVGATAAPGYIAPGYGT 636
Query 93 SHGFDIDAAGFGLCLR 108
+G+ A +GL R
Sbjct 637 PYGYSTAAPAYGLPKR 652
> HsM8922989
Length=490
Score = 27.7 bits (60), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 20/42 (47%), Gaps = 0/42 (0%)
Query 67 GRRLETIRSFVGLADAPGRRALMYAASHGFDIDAAGFGLCLR 108
GR T +FVG APG A Y +G+ A +GL R
Sbjct 429 GRGTLTRGAFVGATAAPGYIAPGYGTPYGYSTAAPAYGLPKR 470
> CE13116
Length=71
Score = 27.3 bits (59), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 16/61 (26%), Positives = 26/61 (42%), Gaps = 0/61 (0%)
Query 16 YKLADKSLCIFLDYDGTLTPIVADPREAKMSPAFRKTLKKLARVFPVAIVTGRRLETIRS 75
+ L DK + F+ D + ++ ++ K P F+ LK L +V A G T
Sbjct 2 FSLHDKDMSGFISKDDVICMLLGAEKDDKKDPVFKTNLKFLIQVIKEADKDGDSKITFDE 61
Query 76 F 76
F
Sbjct 62 F 62
> YFL045c
Length=254
Score = 26.9 bits (58), Expect = 9.0, Method: Compositional matrix adjust.
Identities = 21/55 (38%), Positives = 26/55 (47%), Gaps = 6/55 (10%)
Query 16 YKLADKSLCIFLDYDGTLTPIVADPREAKMSPAFRKTLKKLARVFPVAIVTGRRL 70
YK ++L +F D DGTLTP +S RKTL KL + V G L
Sbjct 8 YKEKPETLVLF-DVDGTLTPA-----RLTVSEEVRKTLAKLRNKCCIGFVGGSDL 56
Lambda K H
0.326 0.141 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1160781780
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40