bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1358_orf1
Length=127
Score E
Sequences producing significant alignments: (Bits) Value
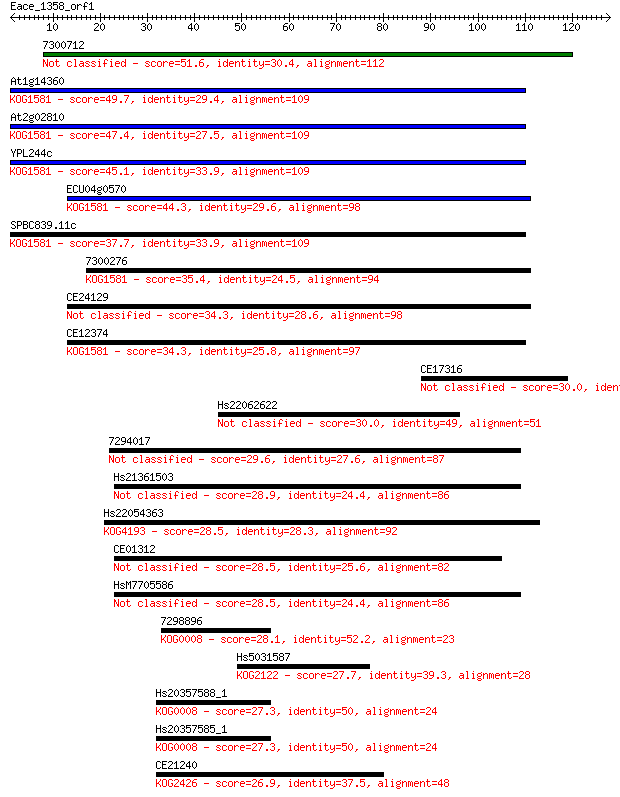
7300712 51.6 4e-07
At1g14360 49.7 1e-06
At2g02810 47.4 7e-06
YPL244c 45.1 4e-05
ECU04g0570 44.3 5e-05
SPBC839.11c 37.7 0.005
7300276 35.4 0.024
CE24129 34.3 0.063
CE12374 34.3 0.065
CE17316 30.0 1.0
Hs22062622 30.0 1.1
7294017 29.6 1.3
Hs21361503 28.9 2.6
Hs22054363 28.5 2.9
CE01312 28.5 3.1
HsM7705586 28.5 3.3
7298896 28.1 4.2
Hs5031587 27.7 5.3
Hs20357588_1 27.3 6.8
Hs20357585_1 27.3 6.8
CE21240 26.9 8.9
> 7300712
Length=338
Score = 51.6 bits (122), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 34/112 (30%), Positives = 55/112 (49%), Gaps = 3/112 (2%)
Query 8 EEKVIKGEAATNSFLGVLMLCVSLLCDGLTGPRQDKLLEREKHLNPLLLMLLTNACGVLW 67
+E + A + LG ++L +SL DGLTG Q+++ ++ + ++
Sbjct 156 KEGKVSNLPAETTLLGEVLLFLSLSMDGLTGAVQERIRAASAPSGQQMMRAMNFWSTLML 215
Query 68 CAAAVAAAEGIRPLYFFLQQPDACGYLFAFAVCGSLGQLFIYQRHLFVLSYG 119
A V E +YF ++ P+A +L AVCG LGQ FI+ L V S+G
Sbjct 216 GVAMVFTGEAKEFMYFTIRHPEAWTHLSLIAVCGVLGQFFIF---LMVASFG 264
> At1g14360
Length=331
Score = 49.7 bits (117), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 32/112 (28%), Positives = 50/112 (44%), Gaps = 3/112 (2%)
Query 1 VFQLCKNEEKVIKGEAATNSFLGVLMLCVSLLCDGLTGPRQDKLLEREKHLNPLLLMLLT 60
+F L K K I A N+ LG + ++L DG T QD + R N +ML
Sbjct 151 MFALLKTSSKTISKLAHPNAPLGYGLCFLNLAFDGFTNATQDSITARYPKTNAWDIMLGM 210
Query 61 NACGVLWCAA---AVAAAEGIRPLYFFLQQPDACGYLFAFAVCGSLGQLFIY 109
N G ++ + G + F Q P+A + + +CG++GQ FI+
Sbjct 211 NLWGTIYNMVYMFGLPHGSGFEAVQFCKQHPEAAWDILMYCLCGAVGQNFIF 262
> At2g02810
Length=332
Score = 47.4 bits (111), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 30/112 (26%), Positives = 49/112 (43%), Gaps = 3/112 (2%)
Query 1 VFQLCKNEEKVIKGEAATNSFLGVLMLCVSLLCDGLTGPRQDKLLEREKHLNPLLLMLLT 60
+F L K K I A N+ LG + ++L DG T QD + R +ML
Sbjct 151 IFALLKTSSKTISKLAHPNAPLGYALCSLNLAFDGFTNATQDSIASRYPKTEAWDIMLGM 210
Query 61 NACGVLWCAA---AVAAAEGIRPLYFFLQQPDACGYLFAFAVCGSLGQLFIY 109
N G ++ + G + + F P+A + + +CG++GQ FI+
Sbjct 211 NLWGTIYNMIYMFGLPQGIGFKAIQFCKLHPEAAWDILKYCICGAVGQNFIF 262
> YPL244c
Length=339
Score = 45.1 bits (105), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 37/124 (29%), Positives = 55/124 (44%), Gaps = 15/124 (12%)
Query 1 VFQLCKNEEKVIK---GEAAT-NSFLGVLMLCVSLLCDGLTGPRQDKLLE----REKHLN 52
+F + N+ K +K E+ N G +L SL DGLT QDKLL+ +EK
Sbjct 149 IFTIGGNDGKKLKRSFNESGNDNKLQGFGLLFSSLFLDGLTNATQDKLLKANKAKEKGKQ 208
Query 53 PLL----LMLLTNACGVLWCA---AAVAAAEGIRPLYFFLQQPDACGYLFAFAVCGSLGQ 105
L+ LM N +LW + + + P GYL ++ CG++GQ
Sbjct 209 TLITGAHLMFTLNLFVILWNILYFIVIDCKQWDNAVSVLTMDPQVWGYLMLYSFCGAMGQ 268
Query 106 LFIY 109
FI+
Sbjct 269 CFIF 272
> ECU04g0570
Length=318
Score = 44.3 bits (103), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 29/100 (29%), Positives = 47/100 (47%), Gaps = 3/100 (3%)
Query 13 KGEAATN-SFLGVLMLCVSLLCDGLTGPRQDKLLEREKHLNPLLLMLLTNACGVLWCAAA 71
KG +A+ S +G+L+L SLL DG QD + R ++ +M +N L A
Sbjct 153 KGSSASGFSIIGILVLITSLLADGAINSSQDHIF-RNFKVSSFHMMYYSNLFRFLISFTA 211
Query 72 VAAAEGIR-PLYFFLQQPDACGYLFAFAVCGSLGQLFIYQ 110
+ + +R + F P+ LF ++ LGQ+ IY
Sbjct 212 ILLTDNLRYSIAFIKSTPEVAPDLFLYSTFNILGQVVIYS 251
> SPBC839.11c
Length=322
Score = 37.7 bits (86), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 37/113 (32%), Positives = 59/113 (52%), Gaps = 9/113 (7%)
Query 1 VFQLCKNEEKVIKGEAAT-NSFLGVLMLCVSLLCDGLTGPRQDKLLEREKHLNPLLLMLL 59
+F +N KG+ A +S +G+L+L +LL DG+T QDK+ + K L+ + +M+
Sbjct 146 IFSYFQNTSS--KGKHAEHDSPIGLLLLFFNLLMDGITNTTQDKVFGKYK-LSSVTMMIA 202
Query 60 TN---ACGVLWCAAAVAAAEGIRPLYFFLQQPDACGYLFAFAVCGSLGQLFIY 109
N AC L ++ +PL F + P + FA GS+GQLFI+
Sbjct 203 VNLGIAC--LNGLYLISPFCNQQPLSFINRHPSILKDMLLFACTGSVGQLFIF 253
> 7300276
Length=465
Score = 35.4 bits (80), Expect = 0.024, Method: Composition-based stats.
Identities = 23/95 (24%), Positives = 43/95 (45%), Gaps = 2/95 (2%)
Query 17 ATNSFLGVLMLCVSLLCDGLTGPRQDKLLEREKHLNPLLLMLLTNACGVLWCAAAVAAAE 76
+ G+ +L + ++ D T Q L + + PL +M N ++ A+++
Sbjct 298 GVTTLTGIFLLSMYMVFDSFTANWQGSLF-KSYGMTPLQMMCGVNLFSSIFTGASLSMQG 356
Query 77 G-IRPLYFFLQQPDACGYLFAFAVCGSLGQLFIYQ 110
G + L F + P + +VC ++GQLFIY
Sbjct 357 GFMDSLAFATEHPKFVFDMVVLSVCSAVGQLFIYH 391
> CE24129
Length=340
Score = 34.3 bits (77), Expect = 0.063, Method: Compositional matrix adjust.
Identities = 28/100 (28%), Positives = 45/100 (45%), Gaps = 3/100 (3%)
Query 13 KGEAATNSF-LGVLMLCVSLLCDGLTGPRQDKLLEREKHLNPLLLMLLTNACGVLWCAAA 71
KG A F G L+L SL DG T QD+ +++ +M TN L+ +A
Sbjct 168 KGGAEDKDFGFGELLLIFSLAMDGTTTSIQDR-IKKSYQRTGTSMMFYTNLYSSLYLSAG 226
Query 72 VAAAEGIRPLYFFLQQ-PDACGYLFAFAVCGSLGQLFIYQ 110
+ + ++F+Q+ P L A+ LGQ I++
Sbjct 227 LLVTGELWSFFYFVQRHPYVFWDLTGLAIASCLGQWCIFK 266
> CE12374
Length=417
Score = 34.3 bits (77), Expect = 0.065, Method: Composition-based stats.
Identities = 25/101 (24%), Positives = 47/101 (46%), Gaps = 4/101 (3%)
Query 13 KGEAAT---NSFLGVLMLCVSLLCDGLTGPRQDKLLEREKHLNPLLLMLLTNACGVLWCA 69
KG +T SF G++++ LL D T Q L + + ++ +M N + CA
Sbjct 257 KGAGSTITYTSFSGMILMAGYLLFDAFTLNWQKALFDTKPKVSKYQMMFGVNFFSAILCA 316
Query 70 AAVAAAEGI-RPLYFFLQQPDACGYLFAFAVCGSLGQLFIY 109
++ + + F + D +F ++ G++GQ+FIY
Sbjct 317 VSLIEQGTLWSSIKFGAEHVDFSRDVFLLSLSGAIGQIFIY 357
> CE17316
Length=308
Score = 30.0 bits (66), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 13/31 (41%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 88 PDACGYLFAFAVCGSLGQLFIYQRHLFVLSY 118
PD YL+AF L +F+Y H F+LS+
Sbjct 129 PDFIWYLYAFFFVNELIGIFLYWAHWFILSF 159
> Hs22062622
Length=418
Score = 30.0 bits (66), Expect = 1.1, Method: Composition-based stats.
Identities = 25/66 (37%), Positives = 28/66 (42%), Gaps = 17/66 (25%)
Query 45 LEREKHLNPLLLMLLTNACG--VLWC-------------AAAVAAAEGIRPLYFFLQQPD 89
L RE H LLL LL +ACG V WC AA A G P Y P
Sbjct 108 LCRESHWKCLLLTLLIHACGAVVAWCRLATVPRLVLGPEAALARGAGGPPPTY--PASPC 165
Query 90 ACGYLF 95
+ GYL+
Sbjct 166 SDGYLY 171
> 7294017
Length=386
Score = 29.6 bits (65), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 24/88 (27%), Positives = 37/88 (42%), Gaps = 1/88 (1%)
Query 22 LGVLMLCVSLLCDGLTGPRQDKLLEREKHLNPLLLMLLTNACGVLWCAAAVAAAEGIRPL 81
LGV M+ +LLCD G Q+K + K + ++ V +
Sbjct 210 LGVAMISGALLCDAAIGNVQEKAMREFKAPSSEVVFYSYGLGFVYLFVIMLVTGNFFSGF 269
Query 82 YFFLQQP-DACGYLFAFAVCGSLGQLFI 108
F L+ P + GY F F++ G LG F+
Sbjct 270 AFCLEHPVETFGYGFLFSLSGYLGIQFV 297
> Hs21361503
Length=401
Score = 28.9 bits (63), Expect = 2.6, Method: Composition-based stats.
Identities = 21/88 (23%), Positives = 43/88 (48%), Gaps = 3/88 (3%)
Query 23 GVLMLCVSLLCDGLTGPRQDKLLEREKHLNPLLLMLLTNACGVLWCAAAVAAAEGIRP-L 81
GV+++ ++L D + G Q+K ++ N ++ L + + G ++ + G+ P +
Sbjct 228 GVVLISLALCADAVIGNVQEKAMKLHNASNSEMV-LYSYSIGFVYILLGLTCTSGLGPAV 286
Query 82 YFFLQQP-DACGYLFAFAVCGSLGQLFI 108
F + P GY F F++ G G F+
Sbjct 287 TFCAKNPVRTYGYAFLFSLTGYFGISFV 314
> Hs22054363
Length=375
Score = 28.5 bits (62), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 26/94 (27%), Positives = 40/94 (42%), Gaps = 15/94 (15%)
Query 21 FLGVLMLCVSL--LCDGLTGPRQDKLLEREKHLNPLLLMLLTNACGVLWCAAAVAAAEGI 78
FL + M V + +C G G R ++ L E N ++ LT G+ W A
Sbjct 188 FLNIAMFIVVMVQIC-GRNGKRSNRTLREEVLRNLRSVVSLTFLLGMTWGFA-------- 238
Query 79 RPLYFFLQQPDACGYLFAFAVCGSLGQLFIYQRH 112
FF P +++ F++ SL LFI+ H
Sbjct 239 ----FFAWGPLNIPFMYLFSIFNSLQGLFIFIFH 268
> CE01312
Length=364
Score = 28.5 bits (62), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 21/83 (25%), Positives = 37/83 (44%), Gaps = 1/83 (1%)
Query 23 GVLMLCVSLLCDGLTGPRQDKLLEREKHLNPLLLMLLTNACGVLWCAAAVAAAEGIRPLY 82
G +M+C +LL D + G Q+K +++ + +++ V A V + E +
Sbjct 189 GYIMICGALLADAVIGNIQEKNMKKYGGSSNEMVLYSYGIGSVFIFAFVVLSGEVFSAIP 248
Query 83 FFLQQP-DACGYLFAFAVCGSLG 104
FFL+ GY + G LG
Sbjct 249 FFLENSWKTFGYALILSCLGYLG 271
> HsM7705586
Length=382
Score = 28.5 bits (62), Expect = 3.3, Method: Composition-based stats.
Identities = 21/88 (23%), Positives = 43/88 (48%), Gaps = 3/88 (3%)
Query 23 GVLMLCVSLLCDGLTGPRQDKLLEREKHLNPLLLMLLTNACGVLWCAAAVAAAEGIRP-L 81
GV+++ ++L D + G Q+K ++ N ++ L + + G ++ + G+ P +
Sbjct 228 GVVLISLALCADAVIGNVQEKAMKLHNASNSEMV-LYSYSIGFVYILLGLTCTSGLGPAV 286
Query 82 YFFLQQP-DACGYLFAFAVCGSLGQLFI 108
F + P GY F F++ G G F+
Sbjct 287 TFCAKNPVRTYGYAFLFSLTGYFGISFV 314
> 7298896
Length=2065
Score = 28.1 bits (61), Expect = 4.2, Method: Composition-based stats.
Identities = 12/23 (52%), Positives = 15/23 (65%), Gaps = 0/23 (0%)
Query 33 CDGLTGPRQDKLLEREKHLNPLL 55
C L R+DKL+ EK +NPLL
Sbjct 1567 CFELLAEREDKLMRLEKAINPLL 1589
> Hs5031587
Length=2303
Score = 27.7 bits (60), Expect = 5.3, Method: Composition-based stats.
Identities = 11/28 (39%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 49 KHLNPLLLMLLTNACGVLWCAAAVAAAE 76
+HL L +++NACG LW +A +A +
Sbjct 632 QHLTSHSLTIVSNACGTLWNLSARSARD 659
> Hs20357588_1
Length=1689
Score = 27.3 bits (59), Expect = 6.8, Method: Composition-based stats.
Identities = 12/24 (50%), Positives = 15/24 (62%), Gaps = 0/24 (0%)
Query 32 LCDGLTGPRQDKLLEREKHLNPLL 55
LCD ++DKL EK +NPLL
Sbjct 1476 LCDEKLKEKEDKLARLEKAINPLL 1499
> Hs20357585_1
Length=1710
Score = 27.3 bits (59), Expect = 6.8, Method: Composition-based stats.
Identities = 12/24 (50%), Positives = 15/24 (62%), Gaps = 0/24 (0%)
Query 32 LCDGLTGPRQDKLLEREKHLNPLL 55
LCD ++DKL EK +NPLL
Sbjct 1497 LCDEKLKEKEDKLARLEKAINPLL 1520
> CE21240
Length=578
Score = 26.9 bits (58), Expect = 8.9, Method: Composition-based stats.
Identities = 18/48 (37%), Positives = 21/48 (43%), Gaps = 4/48 (8%)
Query 32 LCDGLTGPRQDKLLEREKHLNPLLLMLLTNACGVLWCAAAVAAAEGIR 79
L + G D +LE EKHLN L CG + AAE IR
Sbjct 368 LVEACVGGVVDAMLESEKHLNQLDAYAGDGDCG----STFAGAAEAIR 411
Lambda K H
0.330 0.143 0.459
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1176738752
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40