bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1362_orf1
Length=108
Score E
Sequences producing significant alignments: (Bits) Value
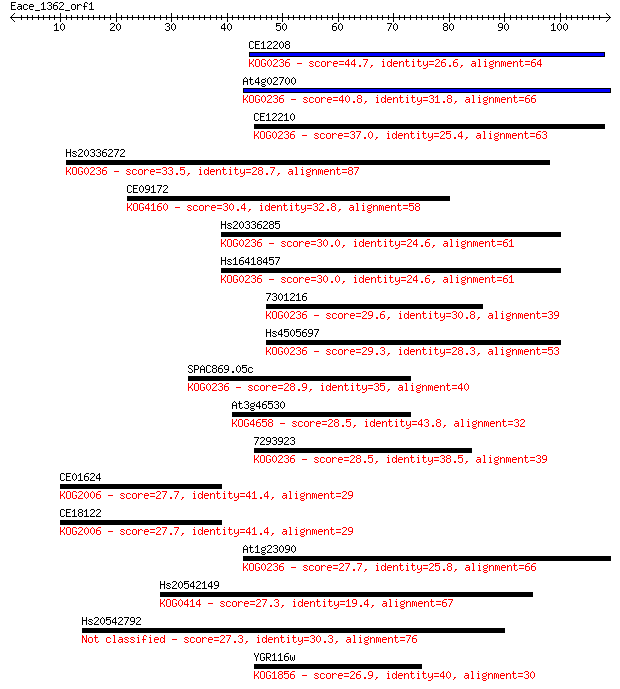
CE12208 44.7 4e-05
At4g02700 40.8 6e-04
CE12210 37.0 0.008
Hs20336272 33.5 0.11
CE09172 30.4 0.78
Hs20336285 30.0 1.0
Hs16418457 30.0 1.0
7301216 29.6 1.4
Hs4505697 29.3 1.9
SPAC869.05c 28.9 2.7
At3g46530 28.5 2.8
7293923 28.5 3.5
CE01624 27.7 4.9
CE18122 27.7 4.9
At1g23090 27.7 5.0
Hs20542149 27.3 6.3
Hs20542792 27.3 6.4
YGR116w 26.9 8.1
> CE12208
Length=749
Score = 44.7 bits (104), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 17/64 (26%), Positives = 37/64 (57%), Gaps = 0/64 (0%)
Query 44 TRVMIVDGSSINDLDVTAIRMLERLVKTLGERGVLLLFANWKGPMRDFLQKAAFYDVLPP 103
+R ++D S +D+ + L+ + + +RG+L+ FAN K P+R+ +K F++ +
Sbjct 646 SRHFVIDCSGFTFIDLMGVSALKEIFSDMRKRGILVYFANAKAPVREMFEKCHFFNFVSK 705
Query 104 EHCF 107
E+ +
Sbjct 706 ENFY 709
> At4g02700
Length=646
Score = 40.8 bits (94), Expect = 6e-04, Method: Composition-based stats.
Identities = 21/66 (31%), Positives = 37/66 (56%), Gaps = 0/66 (0%)
Query 43 STRVMIVDGSSINDLDVTAIRMLERLVKTLGERGVLLLFANWKGPMRDFLQKAAFYDVLP 102
S + +++D S++ ++D + I MLE L K LG R + L+ AN + L K+ F + +
Sbjct 553 SLQYIVLDMSAVGNIDTSGISMLEELNKILGRRELKLVIANPGAEVMKKLSKSTFIESIG 612
Query 103 PEHCFL 108
E +L
Sbjct 613 KERIYL 618
> CE12210
Length=700
Score = 37.0 bits (84), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 16/63 (25%), Positives = 35/63 (55%), Gaps = 0/63 (0%)
Query 45 RVMIVDGSSINDLDVTAIRMLERLVKTLGERGVLLLFANWKGPMRDFLQKAAFYDVLPPE 104
R ++D S +D+ + L+ + L ++ V + FA+ K P+R+ +K +F+D + E
Sbjct 622 RHFVIDCSGFTFIDLMGVSALKEVFSDLRKKRVQVYFASTKVPVREMFEKCSFFDFVSKE 681
Query 105 HCF 107
+ +
Sbjct 682 NFY 684
> Hs20336272
Length=701
Score = 33.5 bits (75), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 25/98 (25%), Positives = 37/98 (37%), Gaps = 11/98 (11%)
Query 11 DAGLGREEEKPGGVQAAVH-----------PVFESQVLHSLLPSTRVMIVDGSSINDLDV 59
DAG K GG + V PV L +++D + + LD
Sbjct 565 DAGCMAARRKEGGSETGVGEGGPAQGEDLGPVSTRAALVPAAAGFHTVVIDCAPLLFLDA 624
Query 60 TAIRMLERLVKTLGERGVLLLFANWKGPMRDFLQKAAF 97
+ L+ L + G G+ LL A P+RD L + F
Sbjct 625 AGVSTLQDLRRDYGALGISLLLACCSPPVRDILSRGGF 662
> CE09172
Length=486
Score = 30.4 bits (67), Expect = 0.78, Method: Compositional matrix adjust.
Identities = 19/58 (32%), Positives = 30/58 (51%), Gaps = 1/58 (1%)
Query 22 GGVQAAVHPVFESQVLHSLLPSTRVMIVDGSSINDLDVTAIRMLERLVKTLGERGVLL 79
GG+ V P +QV LL T++ VD S +DL + LE+L+ + + G+ L
Sbjct 381 GGLNMYVRPSKPNQVKQQLL-ETKINDVDQDSFSDLGLFGAEFLEKLLTEILQMGIAL 437
> Hs20336285
Length=865
Score = 30.0 bits (66), Expect = 1.0, Method: Composition-based stats.
Identities = 15/61 (24%), Positives = 30/61 (49%), Gaps = 0/61 (0%)
Query 39 SLLPSTRVMIVDGSSINDLDVTAIRMLERLVKTLGERGVLLLFANWKGPMRDFLQKAAFY 98
SLLPS +I+D S ++ +D + +L ++ +L+L A + ++ F+
Sbjct 611 SLLPSVHTIILDFSMVHYVDSRGLVVLRQICNAFQNANILILIAGCHSSIVRAFERNDFF 670
Query 99 D 99
D
Sbjct 671 D 671
> Hs16418457
Length=970
Score = 30.0 bits (66), Expect = 1.0, Method: Composition-based stats.
Identities = 15/61 (24%), Positives = 30/61 (49%), Gaps = 0/61 (0%)
Query 39 SLLPSTRVMIVDGSSINDLDVTAIRMLERLVKTLGERGVLLLFANWKGPMRDFLQKAAFY 98
SLLPS +I+D S ++ +D + +L ++ +L+L A + ++ F+
Sbjct 716 SLLPSVHTIILDFSMVHYVDSRGLVVLRQICNAFQNANILILIAGCHSSIVRAFERNDFF 775
Query 99 D 99
D
Sbjct 776 D 776
> 7301216
Length=654
Score = 29.6 bits (65), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 12/39 (30%), Positives = 21/39 (53%), Gaps = 0/39 (0%)
Query 47 MIVDGSSINDLDVTAIRMLERLVKTLGERGVLLLFANWK 85
+++D S + D T +++ L+ +RG LL F N K
Sbjct 575 VVIDASHVYGADFTTATVIDSLISDFNQRGQLLFFYNLK 613
> Hs4505697
Length=780
Score = 29.3 bits (64), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 15/53 (28%), Positives = 30/53 (56%), Gaps = 0/53 (0%)
Query 47 MIVDGSSINDLDVTAIRMLERLVKTLGERGVLLLFANWKGPMRDFLQKAAFYD 99
+++D +I+ LDV +R L +VK V + FA+ + + + L++ F+D
Sbjct 658 LVLDCGAISFLDVVGVRSLRVIVKEFQRIDVNVYFASLQDYVIEKLEQCGFFD 710
> SPAC869.05c
Length=840
Score = 28.9 bits (63), Expect = 2.7, Method: Composition-based stats.
Identities = 14/40 (35%), Positives = 24/40 (60%), Gaps = 0/40 (0%)
Query 33 ESQVLHSLLPSTRVMIVDGSSINDLDVTAIRMLERLVKTL 72
E+ + L P + +I D S++N+LD TA++ L + K L
Sbjct 643 ENSEIEDLRPLLQAIIFDFSAVNNLDTTAVQSLIDIRKEL 682
> At3g46530
Length=835
Score = 28.5 bits (62), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 14/32 (43%), Positives = 20/32 (62%), Gaps = 0/32 (0%)
Query 41 LPSTRVMIVDGSSINDLDVTAIRMLERLVKTL 72
L R + +DG+SIND D+ AI R ++TL
Sbjct 584 LIHLRYLGIDGNSINDFDIAAIISKLRFLQTL 615
> 7293923
Length=742
Score = 28.5 bits (62), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 15/39 (38%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 45 RVMIVDGSSINDLDVTAIRMLERLVKTLGERGVLLLFAN 83
+V+++D S + +DV R L L K L RG LL A+
Sbjct 665 KVLVLDFSMLGHIDVAGCRTLTDLSKELKVRGARLLLAS 703
> CE01624
Length=1941
Score = 27.7 bits (60), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 12/29 (41%), Positives = 14/29 (48%), Gaps = 0/29 (0%)
Query 10 ADAGLGREEEKPGGVQAAVHPVFESQVLH 38
A GREEE PG V+P E + H
Sbjct 9 ASPTTGREEEIPGSSPEGVYPADEDHIFH 37
> CE18122
Length=1943
Score = 27.7 bits (60), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 12/29 (41%), Positives = 14/29 (48%), Gaps = 0/29 (0%)
Query 10 ADAGLGREEEKPGGVQAAVHPVFESQVLH 38
A GREEE PG V+P E + H
Sbjct 9 ASPTTGREEEIPGSSPEGVYPADEDHIFH 37
> At1g23090
Length=631
Score = 27.7 bits (60), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 17/70 (24%), Positives = 36/70 (51%), Gaps = 7/70 (10%)
Query 43 STRVMIVDGSSINDLDVTAIRMLERLVKTLGERGVLLLFANWKGPMRDFLQKAAFYD--- 99
S + +I++ S+++ +D + + L KT ++ + L+F N P+ + ++K D
Sbjct 546 SLQFLILEMSAVSGVDTNGVSFFKELKKTTAKKDIELVFVN---PLSEVVEKLQRADEQK 602
Query 100 -VLPPEHCFL 108
+ PE FL
Sbjct 603 EFMRPEFLFL 612
> Hs20542149
Length=477
Score = 27.3 bits (59), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 13/67 (19%), Positives = 33/67 (49%), Gaps = 0/67 (0%)
Query 28 VHPVFESQVLHSLLPSTRVMIVDGSSINDLDVTAIRMLERLVKTLGERGVLLLFANWKGP 87
+ P+ ES++L+ +L +++ ++ G + + + L+ L E ++ W+G
Sbjct 18 IRPLEESELLYPILGQSKLFVLSGEWSQSSEEASEEVAVTLIYQLAESPTVICAQIWQGC 77
Query 88 MRDFLQK 94
+ L+K
Sbjct 78 AKQALEK 84
> Hs20542792
Length=300
Score = 27.3 bits (59), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 23/77 (29%), Positives = 36/77 (46%), Gaps = 3/77 (3%)
Query 14 LGREEEKPGGVQAAVHPVF-ESQVLHSLLPSTRVMIVDGSSINDLDVTAIRMLERLVKTL 72
LGR E+ A + VF +S + H ++ TR ++G S +D A L+ +VK
Sbjct 101 LGRYTEEEQKTVALIKAVFGKSAMKHMVILFTRKEELEGQSFHDFIADADVGLKSIVKEC 160
Query 73 GERGVLLLFANWKGPMR 89
G R F+N K +
Sbjct 161 GNR--CCAFSNSKKTSK 175
> YGR116w
Length=1451
Score = 26.9 bits (58), Expect = 8.1, Method: Composition-based stats.
Identities = 12/30 (40%), Positives = 17/30 (56%), Gaps = 0/30 (0%)
Query 45 RVMIVDGSSINDLDVTAIRMLERLVKTLGE 74
+V+IVD NDLD + L+ V+ L E
Sbjct 1320 KVLIVDNQKYNDLDQIIVEYLQNKVRLLNE 1349
Lambda K H
0.323 0.140 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1160781780
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40