bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1395_orf1
Length=118
Score E
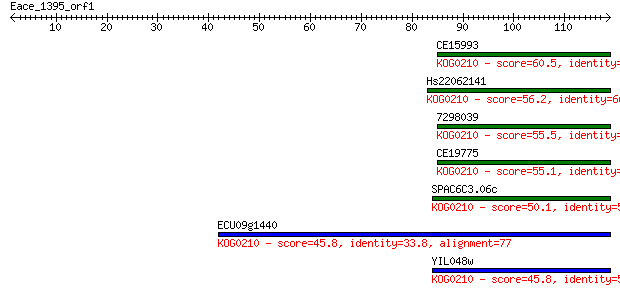
Sequences producing significant alignments: (Bits) Value
CE15993 60.5 8e-10
Hs22062141 56.2 1e-08
7298039 55.5 2e-08
CE19775 55.1 3e-08
SPAC6C3.06c 50.1 9e-07
ECU09g1440 45.8 2e-05
YIL048w 45.8 2e-05
> CE15993
Length=1059
Score = 60.5 bits (145), Expect = 8e-10, Method: Composition-based stats.
Identities = 26/34 (76%), Positives = 30/34 (88%), Gaps = 0/34 (0%)
Query 85 LGLEQVLWANTVVAAGTVIGIVIYTGRETRSSMN 118
L +E VLWANTVVA+GT +GIV+YTGRETRS MN
Sbjct 288 LNVENVLWANTVVASGTAVGIVVYTGRETRSVMN 321
> Hs22062141
Length=875
Score = 56.2 bits (134), Expect = 1e-08, Method: Composition-based stats.
Identities = 24/36 (66%), Positives = 29/36 (80%), Gaps = 0/36 (0%)
Query 83 EPLGLEQVLWANTVVAAGTVIGIVIYTGRETRSSMN 118
E L +E LWA TVVA+GTV+G+V+YTGRE RS MN
Sbjct 78 ESLSIENTLWAGTVVASGTVVGVVLYTGRELRSVMN 113
> 7298039
Length=1058
Score = 55.5 bits (132), Expect = 2e-08, Method: Composition-based stats.
Identities = 26/34 (76%), Positives = 27/34 (79%), Gaps = 0/34 (0%)
Query 85 LGLEQVLWANTVVAAGTVIGIVIYTGRETRSSMN 118
L +E LWANTVVAAGT GIVIYTG ETRS MN
Sbjct 259 LSVENTLWANTVVAAGTATGIVIYTGCETRSVMN 292
> CE19775
Length=1064
Score = 55.1 bits (131), Expect = 3e-08, Method: Composition-based stats.
Identities = 24/34 (70%), Positives = 29/34 (85%), Gaps = 0/34 (0%)
Query 85 LGLEQVLWANTVVAAGTVIGIVIYTGRETRSSMN 118
L +E VLWANTVV++GT IG+V+YTG ETRS MN
Sbjct 293 LDVENVLWANTVVSSGTAIGVVVYTGCETRSVMN 326
> SPAC6C3.06c
Length=1033
Score = 50.1 bits (118), Expect = 9e-07, Method: Composition-based stats.
Identities = 19/35 (54%), Positives = 28/35 (80%), Gaps = 0/35 (0%)
Query 84 PLGLEQVLWANTVVAAGTVIGIVIYTGRETRSSMN 118
P+ ++ LWANTV+A+ V G+V+YTG++TR SMN
Sbjct 268 PISVDHTLWANTVLASDGVYGVVVYTGKDTRQSMN 302
> ECU09g1440
Length=1094
Score = 45.8 bits (107), Expect = 2e-05, Method: Composition-based stats.
Identities = 26/78 (33%), Positives = 40/78 (51%), Gaps = 1/78 (1%)
Query 42 SSGAATPAAGTA-ATAAAAAAAAGPGDTMVDVSGGGCSYCAVEPLGLEQVLWANTVVAAG 100
S G + AAG++ + G DVS E LG E +LW NTVVA
Sbjct 278 SDGRDSAAAGSSEGLEVGNESNVGRETEEADVSLKDEEKPVAEALGCENMLWMNTVVATC 337
Query 101 TVIGIVIYTGRETRSSMN 118
+++G V+YTG++ ++ +N
Sbjct 338 SILGCVVYTGKDAKAMLN 355
> YIL048w
Length=1151
Score = 45.8 bits (107), Expect = 2e-05, Method: Composition-based stats.
Identities = 20/36 (55%), Positives = 27/36 (75%), Gaps = 1/36 (2%)
Query 84 PLGLEQVLWANTVVAA-GTVIGIVIYTGRETRSSMN 118
PL ++ LWANTV+A+ G I V+YTGR+TR +MN
Sbjct 361 PLSVDNTLWANTVLASSGFCIACVVYTGRDTRQAMN 396
Lambda K H
0.309 0.121 0.338
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1167969826
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40