bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
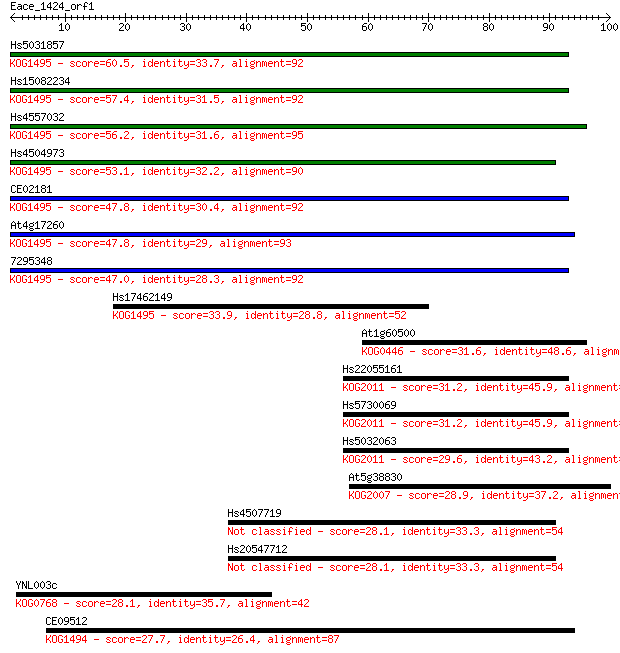
Query= Eace_1424_orf1
Length=99
Score E
Sequences producing significant alignments: (Bits) Value
Hs5031857 60.5 7e-10
Hs15082234 57.4 6e-09
Hs4557032 56.2 1e-08
Hs4504973 53.1 1e-07
CE02181 47.8 5e-06
At4g17260 47.8 6e-06
7295348 47.0 9e-06
Hs17462149 33.9 0.077
At1g60500 31.6 0.39
Hs22055161 31.2 0.46
Hs5730069 31.2 0.48
Hs5032063 29.6 1.3
At5g38830 28.9 2.2
Hs4507719 28.1 4.7
Hs20547712 28.1 4.7
YNL003c 28.1 4.7
CE09512 27.7 5.5
> Hs5031857
Length=332
Score = 60.5 bits (145), Expect = 7e-10, Method: Composition-based stats.
Identities = 31/93 (33%), Positives = 58/93 (62%), Gaps = 3/93 (3%)
Query 1 EIVRLLGQGSAYYAPGASAIQMAESYLKDRKRVMVCSCYLQGQYGVQNH-YLGVPCVIGG 59
E+++L G S +A G S +AES +K+ +RV S ++G YG+++ +L VPC++G
Sbjct 240 EVIKLKGYTS--WAIGLSVADLAESIMKNLRRVHPVSTMIKGLYGIKDDVFLSVPCILGQ 297
Query 60 RGVEKIIELELTAQERQELQGSIDEVKEMQKAI 92
G+ ++++ LT++E L+ S D + +QK +
Sbjct 298 NGISDLVKVTLTSEEEARLKKSADTLWGIQKEL 330
> Hs15082234
Length=381
Score = 57.4 bits (137), Expect = 6e-09, Method: Composition-based stats.
Identities = 29/93 (31%), Positives = 54/93 (58%), Gaps = 3/93 (3%)
Query 1 EIVRLLGQGSAYYAPGASAIQMAESYLKDRKRVMVCSCYLQGQYGVQNH-YLGVPCVIGG 59
EI+++ G S +A G S + ES LK+ +R+ S ++G YG+ +L +PC++G
Sbjct 289 EIIKMKGYTS--WAIGLSVADLTESILKNLRRIHPVSTIIKGLYGIDEEVFLSIPCILGE 346
Query 60 RGVEKIIELELTAQERQELQGSIDEVKEMQKAI 92
G+ +I+++LT +E L+ S + E+Q +
Sbjct 347 NGITNLIKIKLTPEEEAHLKKSAKTLWEIQNKL 379
> Hs4557032
Length=334
Score = 56.2 bits (134), Expect = 1e-08, Method: Composition-based stats.
Identities = 30/96 (31%), Positives = 55/96 (57%), Gaps = 3/96 (3%)
Query 1 EIVRLLGQGSAYYAPGASAIQMAESYLKDRKRVMVCSCYLQGQYGVQNH-YLGVPCVIGG 59
E+++L +G +A G S + ES LK+ R+ S ++G YG++N +L +PC++
Sbjct 241 EVIKL--KGYTNWAIGLSVADLIESMLKNLSRIHPVSTMVKGMYGIENEVFLSLPCILNA 298
Query 60 RGVEKIIELELTAQERQELQGSIDEVKEMQKAIAAL 95
RG+ +I +L E +L+ S D + ++QK + L
Sbjct 299 RGLTSVINQKLKDDEVAQLKKSADTLWDIQKDLKDL 334
> Hs4504973
Length=332
Score = 53.1 bits (126), Expect = 1e-07, Method: Composition-based stats.
Identities = 29/91 (31%), Positives = 53/91 (58%), Gaps = 3/91 (3%)
Query 1 EIVRLLGQGSAYYAPGASAIQMAESYLKDRKRVMVCSCYLQGQYGVQNH-YLGVPCVIGG 59
EI++L G S +A G S + + S LK+ +RV S ++G YG++ +L +PCV+G
Sbjct 240 EIIKLKGYTS--WAIGLSVMDLVGSILKNLRRVHPVSTMVKGLYGIKEELFLSIPCVLGR 297
Query 60 RGVEKIIELELTAQERQELQGSIDEVKEMQK 90
GV ++++ L ++E + S + + +QK
Sbjct 298 NGVSDVVKINLNSEEEALFKKSAETLWNIQK 328
> CE02181
Length=333
Score = 47.8 bits (112), Expect = 5e-06, Method: Composition-based stats.
Identities = 28/93 (30%), Positives = 51/93 (54%), Gaps = 3/93 (3%)
Query 1 EIVRLLGQGSAYYAPGASAIQMAESYLKDRKRVMVCSCYLQGQYGVQNH-YLGVPCVIGG 59
EI++L G S +A G S ++A+ + + V S ++G +G+ + YL +P V+G
Sbjct 241 EIIKLKGYTS--WAIGLSVAKIAQGIFSNSRNVFALSTNVKGFHGINDDVYLSLPVVLGS 298
Query 60 RGVEKIIELELTAQERQELQGSIDEVKEMQKAI 92
G+ +++ +LT E Q+L S + E+Q I
Sbjct 299 AGLTHVVKQQLTEAEVQKLHNSAKALLEVQNGI 331
> At4g17260
Length=353
Score = 47.8 bits (112), Expect = 6e-06, Method: Composition-based stats.
Identities = 27/95 (28%), Positives = 52/95 (54%), Gaps = 4/95 (4%)
Query 1 EIVRLLGQGSAYYAPGASAIQMAESYLKDRKRVMVCSCYLQGQYGVQ--NHYLGVPCVIG 58
E++ L G S +A G S +A + L+D++++ + +G YGV + +L +P ++G
Sbjct 260 EVIGLKGYTS--WAIGYSVANLARTILRDQRKIHPVTVLARGFYGVDGGDVFLSLPALLG 317
Query 59 GRGVEKIIELELTAQERQELQGSIDEVKEMQKAIA 93
GV + + +T +E ++LQ S + EMQ +
Sbjct 318 RNGVVAVTNVHMTDEEAEKLQKSAKTILEMQSQLG 352
> 7295348
Length=332
Score = 47.0 bits (110), Expect = 9e-06, Method: Composition-based stats.
Identities = 26/93 (27%), Positives = 53/93 (56%), Gaps = 3/93 (3%)
Query 1 EIVRLLGQGSAYYAPGASAIQMAESYLKDRKRVMVCSCYLQGQYGV-QNHYLGVPCVIGG 59
E+++L G S +A G S +A + L++ V S + G++G+ ++ +L +PCV+
Sbjct 240 EVIKLKGYTS--WAIGLSTASLASAILRNTSSVAAVSTSVLGEHGIDKDVFLSLPCVLNA 297
Query 60 RGVEKIIELELTAQERQELQGSIDEVKEMQKAI 92
GV +++ LT E ++LQ S + + ++Q +
Sbjct 298 NGVTSVVKQILTPTEVEQLQKSANIMSDVQAGL 330
> Hs17462149
Length=326
Score = 33.9 bits (76), Expect = 0.077, Method: Composition-based stats.
Identities = 15/53 (28%), Positives = 33/53 (62%), Gaps = 1/53 (1%)
Query 18 SAIQMAESYLKDRKRVMVCSCYLQGQYGVQNH-YLGVPCVIGGRGVEKIIELE 69
S +AES +K+ +RV S ++G G+++ +L V C++G G+ +++++
Sbjct 64 SGADLAESIMKNLRRVHPISTMIKGFCGIKDDVFLSVACILGQNGISDLVKVK 116
> At1g60500
Length=669
Score = 31.6 bits (70), Expect = 0.39, Method: Compositional matrix adjust.
Identities = 18/38 (47%), Positives = 27/38 (71%), Gaps = 1/38 (2%)
Query 59 GRGVEKIIE-LELTAQERQELQGSIDEVKEMQKAIAAL 95
G GVEK++E L A +R++LQ SI +KE + A+AA+
Sbjct 627 GGGVEKMLEESPLVASKREKLQNSIKLLKESKDAVAAI 664
> Hs22055161
Length=1268
Score = 31.2 bits (69), Expect = 0.46, Method: Composition-based stats.
Identities = 17/37 (45%), Positives = 24/37 (64%), Gaps = 3/37 (8%)
Query 56 VIGGRGVEKIIELELTAQERQELQGSIDEVKEMQKAI 92
+IG R E+ LEL Q+R+ELQ + DE++ M AI
Sbjct 255 MIGKRANER---LELLLQKRKELQENQDEIENMMNAI 288
> Hs5730069
Length=1162
Score = 31.2 bits (69), Expect = 0.48, Method: Composition-based stats.
Identities = 17/37 (45%), Positives = 24/37 (64%), Gaps = 3/37 (8%)
Query 56 VIGGRGVEKIIELELTAQERQELQGSIDEVKEMQKAI 92
+IG R E+ LEL Q+R+ELQ + DE++ M AI
Sbjct 186 MIGKRANER---LELLLQKRKELQENQDEIENMMNAI 219
> Hs5032063
Length=1258
Score = 29.6 bits (65), Expect = 1.3, Method: Composition-based stats.
Identities = 16/37 (43%), Positives = 24/37 (64%), Gaps = 3/37 (8%)
Query 56 VIGGRGVEKIIELELTAQERQELQGSIDEVKEMQKAI 92
+IG R E+ LEL Q+R+ELQ + DE++ M +I
Sbjct 258 MIGKRANER---LELLLQKRKELQENQDEIENMMNSI 291
> At5g38830
Length=511
Score = 28.9 bits (63), Expect = 2.2, Method: Composition-based stats.
Identities = 16/45 (35%), Positives = 28/45 (62%), Gaps = 3/45 (6%)
Query 57 IGGRGVEKIIELELTAQERQELQGSIDEVKE--MQKAIAALDASK 99
IG G+ ++IE +TA++ ++ S DE++E +K IA +D K
Sbjct 452 IGEEGISQLIEERITARKNKDFAKS-DEIREKLTRKGIALMDIGK 495
> Hs4507719
Length=841
Score = 28.1 bits (61), Expect = 4.7, Method: Composition-based stats.
Identities = 18/56 (32%), Positives = 27/56 (48%), Gaps = 2/56 (3%)
Query 37 SCYLQGQYGVQNHYLGVPCVIGGRGVEKIIELELTAQERQE--LQGSIDEVKEMQK 90
SC L+ VQN + P V + E II +T + + E L ++E KE Q+
Sbjct 305 SCELRNLKSVQNSHFKEPLVSDEKSSELIITDSITLKNKTESSLLAKLEETKEYQE 360
> Hs20547712
Length=841
Score = 28.1 bits (61), Expect = 4.7, Method: Composition-based stats.
Identities = 18/56 (32%), Positives = 27/56 (48%), Gaps = 2/56 (3%)
Query 37 SCYLQGQYGVQNHYLGVPCVIGGRGVEKIIELELTAQERQE--LQGSIDEVKEMQK 90
SC L+ VQN + P V + E II +T + + E L ++E KE Q+
Sbjct 305 SCELRNLKSVQNSHFKEPLVSDEKSSELIITDSITLKNKTESSLLAKLEETKEYQE 360
> YNL003c
Length=284
Score = 28.1 bits (61), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 15/42 (35%), Positives = 20/42 (47%), Gaps = 0/42 (0%)
Query 2 IVRLLGQGSAYYAPGASAIQMAESYLKDRKRVMVCSCYLQGQ 43
I R LG APGAS ++ Y+K + R + Y QG
Sbjct 47 IYRGLGSAVVASAPGASLFFISYDYMKVKSRPYISKLYSQGS 88
> CE09512
Length=341
Score = 27.7 bits (60), Expect = 5.5, Method: Composition-based stats.
Identities = 23/90 (25%), Positives = 43/90 (47%), Gaps = 3/90 (3%)
Query 7 GQGSAYYAPGASAIQMAESYLKDRKRVMVCSCYLQGQYGVQN-HYLGVPCVIGGRGVEKI 65
G GSA + + + A + ++ K C V+ Y P +G GVEKI
Sbjct 247 GAGSATLSMALAGARFANALVRGIKGEKNVQCAYVASDAVKGVEYFSTPVELGPNGVEKI 306
Query 66 IEL-ELTAQERQELQGSIDEV-KEMQKAIA 93
+ + +++A E++ + S+ E+ K + K +A
Sbjct 307 LGVGKVSAYEQKLIDASVPELNKNIAKGVA 336
Lambda K H
0.317 0.134 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1191270180
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40