bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1441_orf1
Length=152
Score E
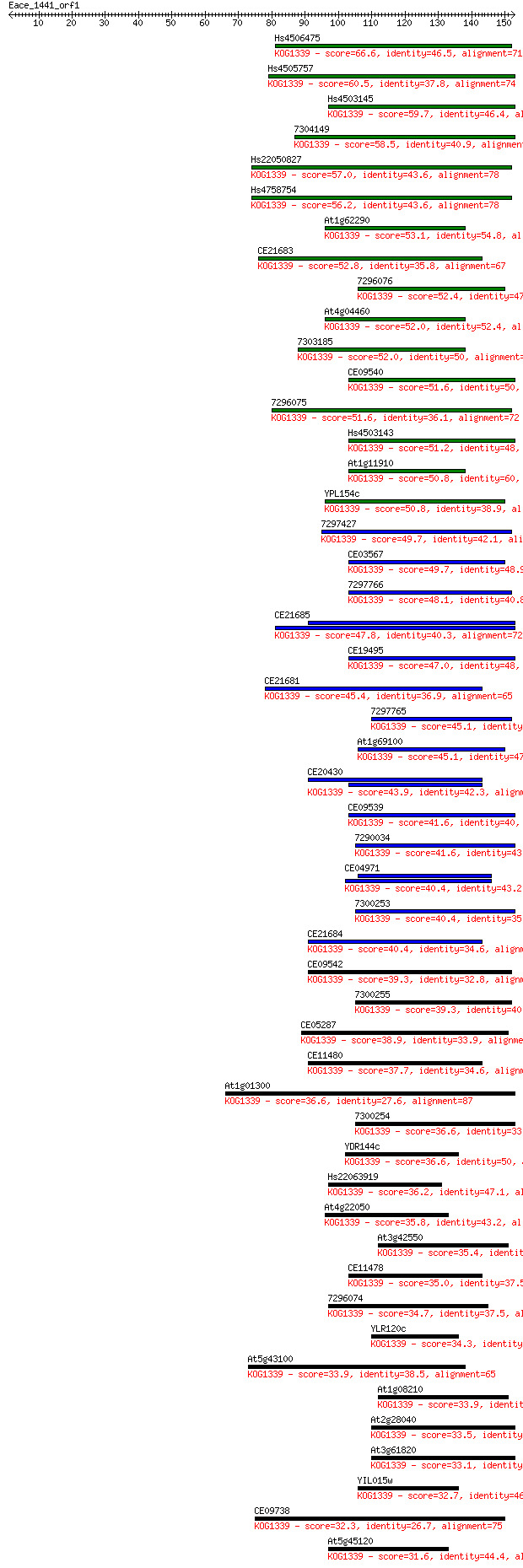
Sequences producing significant alignments: (Bits) Value
Hs4506475 66.6 2e-11
Hs4505757 60.5 1e-09
Hs4503145 59.7 2e-09
7304149 58.5 4e-09
Hs22050827 57.0 1e-08
Hs4758754 56.2 2e-08
At1g62290 53.1 2e-07
CE21683 52.8 3e-07
7296076 52.4 3e-07
At4g04460 52.0 4e-07
7303185 52.0 5e-07
CE09540 51.6 5e-07
7296075 51.6 6e-07
Hs4503143 51.2 8e-07
At1g11910 50.8 9e-07
YPL154c 50.8 1e-06
7297427 49.7 2e-06
CE03567 49.7 2e-06
7297766 48.1 7e-06
CE21685 47.8 7e-06
CE19495 47.0 1e-05
CE21681 45.4 4e-05
7297765 45.1 5e-05
At1g69100 45.1 6e-05
CE20430 43.9 1e-04
CE09539 41.6 6e-04
7290034 41.6 6e-04
CE04971 40.4 0.001
7300253 40.4 0.001
CE21684 40.4 0.001
CE09542 39.3 0.003
7300255 39.3 0.003
CE05287 38.9 0.004
CE11480 37.7 0.008
At1g01300 36.6 0.020
7300254 36.6 0.020
YDR144c 36.6 0.021
Hs22063919 36.2 0.026
At4g22050 35.8 0.031
At3g42550 35.4 0.046
CE11478 35.0 0.057
7296074 34.7 0.070
YLR120c 34.3 0.085
At5g43100 33.9 0.13
At1g08210 33.9 0.14
At2g28040 33.5 0.15
At3g61820 33.1 0.20
YIL015w 32.7 0.30
CE09738 32.3 0.32
At5g45120 31.6 0.67
> Hs4506475
Length=406
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 33/73 (45%), Positives = 45/73 (61%), Gaps = 2/73 (2%)
Query 81 AQPAATLELGLAQTSVPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTD- 139
+QP L LG +SV + D+Q+ G +GIGTPPQ + +FDTGS+N+WV SS+C+
Sbjct 61 SQPMKRLTLGNTTSSVILTNYMDTQYYGEIGIGTPPQTFKVVFDTGSSNVWVPSSKCSRL 120
Query 140 -ETCVKVTRFDPS 151
CV FD S
Sbjct 121 YTACVYHKLFDAS 133
> Hs4505757
Length=388
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 28/74 (37%), Positives = 39/74 (52%), Gaps = 0/74 (0%)
Query 79 HPAQPAATLELGLAQTSVPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCT 138
H PA G + + D+ + G + IGTPPQ +FDTGS+NLWV S C
Sbjct 46 HKYDPAWKYRFGDLSVTYEPMAYMDAAYFGEISIGTPPQNFLVLFDTGSSNLWVPSVYCQ 105
Query 139 DETCVKVTRFDPSK 152
+ C +RF+PS+
Sbjct 106 SQACTSHSRFNPSE 119
> Hs4503145
Length=396
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 26/56 (46%), Positives = 35/56 (62%), Gaps = 0/56 (0%)
Query 97 PILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKVTRFDPSK 152
P++ D ++ G + IG+PPQ IFDTGS+NLWV S CT C +RF PS+
Sbjct 69 PLINYLDMEYFGTISIGSPPQNFTVIFDTGSSNLWVPSVYCTSPACKTHSRFQPSQ 124
> 7304149
Length=392
Score = 58.5 bits (140), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 27/68 (39%), Positives = 40/68 (58%), Gaps = 2/68 (2%)
Query 87 LELGLAQTSVPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRC--TDETCVK 144
+ G P+ D+Q+ G + IG+PPQ R +FDTGS+NLWV S +C T+ C+
Sbjct 54 IRYGGGDVPEPLSNYMDAQYYGPIAIGSPPQNFRVVFDTGSSNLWVPSKKCHLTNIACLM 113
Query 145 VTRFDPSK 152
++D SK
Sbjct 114 HNKYDASK 121
> Hs22050827
Length=160
Score = 57.0 bits (136), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 34/85 (40%), Positives = 47/85 (55%), Gaps = 7/85 (8%)
Query 74 LSFLQHPAQPAATLELGLAQ-----TSVPILQLKDSQFCGMLGIGTPPQWVRPIFDTGST 128
L+ L+ +PA +LG SVP+ + D+Q+ G +G+GTPPQ FDTGS+
Sbjct 41 LNLLRGWGKPAELPKLGAPSPGDKPASVPLSKFLDAQYFGEIGLGTPPQNFTVAFDTGSS 100
Query 129 NLWVVSSRCT--DETCVKVTRFDPS 151
NLWV S RC C RF+P+
Sbjct 101 NLWVPSRRCHFFSVPCWFHHRFNPN 125
> Hs4758754
Length=420
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 34/85 (40%), Positives = 44/85 (51%), Gaps = 7/85 (8%)
Query 74 LSFLQHPAQPAATLELGLAQTS-----VPILQLKDSQFCGMLGIGTPPQWVRPIFDTGST 128
L+ L+ +PA +LG VP+ +D Q+ G +G+GTPPQ FDTGS+
Sbjct 41 LNLLRGWREPAELPKLGAPSPGDKPIFVPLSNYRDVQYFGEIGLGTPPQNFTVAFDTGSS 100
Query 129 NLWVVSSRCT--DETCVKVTRFDPS 151
NLWV S RC C RFDP
Sbjct 101 NLWVPSRRCHFFSVPCWLHHRFDPK 125
> At1g62290
Length=526
Score = 53.1 bits (126), Expect = 2e-07, Method: Composition-based stats.
Identities = 23/42 (54%), Positives = 29/42 (69%), Gaps = 0/42 (0%)
Query 96 VPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRC 137
VP+ D+Q+ G + IGTPPQ IFDTGS+NLWV S +C
Sbjct 79 VPLKNYLDAQYYGEIAIGTPPQKFTVIFDTGSSNLWVPSGKC 120
> CE21683
Length=320
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 24/67 (35%), Positives = 35/67 (52%), Gaps = 0/67 (0%)
Query 76 FLQHPAQPAATLELGLAQTSVPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSS 135
+ + AQ L L Q S P + D + G + +GTP Q + + DTGS+NLWV+ +
Sbjct 39 YQEFLAQQNVARTLQLKQGSQPFIDYMDDFYLGNISVGTPGQTLTLVLDTGSSNLWVIDA 98
Query 136 RCTDETC 142
C E C
Sbjct 99 ACNSEAC 105
> 7296076
Length=405
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 21/46 (45%), Positives = 33/46 (71%), Gaps = 2/46 (4%)
Query 106 FCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRC--TDETCVKVTRFD 149
+ G++GIGTP Q+ + +FDTGS NLWV S++C TD C + +++
Sbjct 93 YYGLIGIGTPEQYFKVVFDTGSANLWVPSAQCLATDVACQQHNQYN 138
> At4g04460
Length=508
Score = 52.0 bits (123), Expect = 4e-07, Method: Composition-based stats.
Identities = 22/42 (52%), Positives = 30/42 (71%), Gaps = 0/42 (0%)
Query 96 VPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRC 137
VP+ D+Q+ G + IGTPPQ IFDTGS+NLW+ S++C
Sbjct 77 VPLKNYLDAQYYGDITIGTPPQKFTVIFDTGSSNLWIPSTKC 118
> 7303185
Length=404
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 25/50 (50%), Positives = 32/50 (64%), Gaps = 0/50 (0%)
Query 88 ELGLAQTSVPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRC 137
E A S P+ D+Q+ G + IGTPPQ + IFDTGS+NLWV S+ C
Sbjct 67 EWNSAVKSTPLSNYLDAQYFGPITIGTPPQTFKVIFDTGSSNLWVPSATC 116
> CE09540
Length=395
Score = 51.6 bits (122), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 25/50 (50%), Positives = 30/50 (60%), Gaps = 1/50 (2%)
Query 103 DSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKVTRFDPSK 152
D ++ G + IGTPPQ + DTGS NLWV S C D TC + FD SK
Sbjct 70 DVEYLGNITIGTPPQQFIVVLDTGSANLWVPGSNC-DGTCKGKSEFDSSK 118
> 7296075
Length=410
Score = 51.6 bits (122), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 26/74 (35%), Positives = 38/74 (51%), Gaps = 13/74 (17%)
Query 80 PAQPAATLELGLAQTSVPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTD 139
P ATLE L ++++ LG G PPQ ++ + DTGS NLWV+SS+C D
Sbjct 75 PGSKVATLE-----------NLYNTEYYTTLGFGNPPQDLKVLIDTGSANLWVLSSKCPD 123
Query 140 ET--CVKVTRFDPS 151
C +++ S
Sbjct 124 SVAPCANRIKYNSS 137
> Hs4503143
Length=412
Score = 51.2 bits (121), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 24/52 (46%), Positives = 32/52 (61%), Gaps = 2/52 (3%)
Query 103 DSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCT--DETCVKVTRFDPSK 152
D+Q+ G +GIGTPPQ +FDTGS+NLWV S C D C +++ K
Sbjct 76 DAQYYGEIGIGTPPQCFTVVFDTGSSNLWVPSIHCKLLDIACWIHHKYNSDK 127
> At1g11910
Length=506
Score = 50.8 bits (120), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 21/35 (60%), Positives = 27/35 (77%), Gaps = 0/35 (0%)
Query 103 DSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRC 137
D+Q+ G + IGTPPQ +FDTGS+NLWV SS+C
Sbjct 79 DAQYYGEIAIGTPPQKFTVVFDTGSSNLWVPSSKC 113
> YPL154c
Length=405
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 21/54 (38%), Positives = 33/54 (61%), Gaps = 0/54 (0%)
Query 96 VPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKVTRFD 149
VP+ ++Q+ + +GTPPQ + I DTGS+NLWV S+ C C +++D
Sbjct 81 VPLTNYLNAQYYTDITLGTPPQNFKVILDTGSSNLWVPSNECGSLACFLHSKYD 134
> 7297427
Length=372
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 24/60 (40%), Positives = 36/60 (60%), Gaps = 3/60 (5%)
Query 95 SVPILQLKDSQ---FCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKVTRFDPS 151
SV QL +S + G + IGTP Q + +FD+GS+NLWV S+ C + C+ ++D S
Sbjct 55 SVDEEQLSNSMNMAYYGAISIGTPAQSFKVLFDSGSSNLWVPSNTCKSDACLTHNQYDSS 114
> CE03567
Length=444
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 23/49 (46%), Positives = 31/49 (63%), Gaps = 2/49 (4%)
Query 103 DSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRC--TDETCVKVTRFD 149
D+Q+ G + IGTP Q IFDTGS+NLW+ S +C D C+ R+D
Sbjct 91 DAQYFGTISIGTPAQNFTVIFDTGSSNLWIPSKKCPFYDIACMLHHRYD 139
> 7297766
Length=391
Score = 48.1 bits (113), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 20/51 (39%), Positives = 33/51 (64%), Gaps = 2/51 (3%)
Query 103 DSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRC--TDETCVKVTRFDPS 151
++++ G++ IGTP Q +FDTGS NLWV S+ C ++ C + ++D S
Sbjct 73 NNEYYGVIAIGTPEQRFNILFDTGSANLWVPSASCPASNTACQRHNKYDSS 123
> CE21685
Length=829
Score = 47.8 bits (112), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 25/69 (36%), Positives = 33/69 (47%), Gaps = 7/69 (10%)
Query 91 LAQTSVPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETC-------V 143
L S PI D + G +GTPPQ V + DTGS N+WV+ + C + C
Sbjct 488 LNTISQPISDYSDEVYLGNFTVGTPPQPVSLVLDTGSANMWVIDASCDNMFCNGWIGSNY 547
Query 144 KVTRFDPSK 152
+FD SK
Sbjct 548 TRQKFDTSK 556
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 25/79 (31%), Positives = 39/79 (49%), Gaps = 8/79 (10%)
Query 81 AQPAATLELGLAQTSVPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDE 140
+Q AA L+ L S P++ D + + +GTPPQ + DT S NLWV+ + C +
Sbjct 44 SQHAARLQ-QLNTGSQPLIDYYDDMYLANITVGTPPQPASVVLDTASANLWVIDAACNSQ 102
Query 141 TC-------VKVTRFDPSK 152
C +F+P+K
Sbjct 103 ACNGNPGSGYTKQKFNPNK 121
> CE19495
Length=398
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 24/52 (46%), Positives = 32/52 (61%), Gaps = 2/52 (3%)
Query 103 DSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRC--TDETCVKVTRFDPSK 152
++Q+ G + IGTPPQ + +FDTGS+NLWV + C D C RFD K
Sbjct 66 NAQYYGPVTIGTPPQNFQVLFDTGSSNLWVPCANCPFGDIACRMHNRFDCKK 117
> CE21681
Length=396
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 24/65 (36%), Positives = 34/65 (52%), Gaps = 5/65 (7%)
Query 78 QHPAQPAATLELGLAQTSVPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRC 137
QH A+ A L G P + D + G + +GTPPQ + DTGS+NLWV+ + C
Sbjct 45 QHAAR-AQQLNTGFQ----PFVDYFDDFYLGNITLGTPPQPATVVLDTGSSNLWVIDAAC 99
Query 138 TDETC 142
+ C
Sbjct 100 KTQAC 104
> 7297765
Length=423
Score = 45.1 bits (105), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 22/44 (50%), Positives = 29/44 (65%), Gaps = 2/44 (4%)
Query 110 LGIGTPPQWVRPIFDTGSTNLWVVSSRC--TDETCVKVTRFDPS 151
L IGTPPQ FDTGS++LWV S +C T+E C K +++ S
Sbjct 78 LCIGTPPQCFNLQFDTGSSDLWVPSVKCSSTNEACQKHNKYNSS 121
> At1g69100
Length=343
Score = 45.1 bits (105), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 21/44 (47%), Positives = 28/44 (63%), Gaps = 0/44 (0%)
Query 106 FCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKVTRFD 149
F G + +G+PPQ +FDTGST+LWV S +ET K +FD
Sbjct 47 FYGEISVGSPPQKFNVVFDTGSTDLWVPSKEWPEETDHKHPKFD 90
> CE20430
Length=638
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 22/52 (42%), Positives = 26/52 (50%), Gaps = 0/52 (0%)
Query 91 LAQTSVPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETC 142
L S P+L D G + IG+PPQ DT S NLWV+ S CT C
Sbjct 47 LNTGSQPLLGFSDDPCIGNITIGSPPQSAFVFMDTTSANLWVMGSACTSVNC 98
Score = 42.0 bits (97), Expect = 4e-04, Method: Composition-based stats.
Identities = 18/40 (45%), Positives = 21/40 (52%), Gaps = 0/40 (0%)
Query 103 DSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETC 142
D G + IGTPPQ DT S N WV+ S+CT C
Sbjct 451 DDSCIGNITIGTPPQSASVFMDTTSANWWVIGSKCTSANC 490
> CE09539
Length=393
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 20/50 (40%), Positives = 28/50 (56%), Gaps = 1/50 (2%)
Query 103 DSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKVTRFDPSK 152
D ++ G + IGTPPQ + DTGS+NLWV C D +C + +K
Sbjct 70 DFEYLGNITIGTPPQPFLVVLDTGSSNLWVPGPSC-DGSCKGKREYQSTK 118
> 7290034
Length=407
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 21/48 (43%), Positives = 25/48 (52%), Gaps = 0/48 (0%)
Query 105 QFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKVTRFDPSK 152
Q+ G + IGTP Q FDTGS+NLWV S C C F +K
Sbjct 76 QYYGNISIGTPGQDFLVQFDTGSSNLWVPGSSCISTACQDHQVFYKNK 123
> CE04971
Length=709
Score = 40.4 bits (93), Expect = 0.001, Method: Composition-based stats.
Identities = 18/42 (42%), Positives = 26/42 (61%), Gaps = 2/42 (4%)
Query 106 FCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDE--TCVKV 145
+ G + +G+PPQ R I DTGS+N W+ S C + +C KV
Sbjct 40 YVGKITVGSPPQEFRVIMDTGSSNFWIPDSTCDTKATSCHKV 81
Score = 38.5 bits (88), Expect = 0.005, Method: Composition-based stats.
Identities = 19/46 (41%), Positives = 25/46 (54%), Gaps = 2/46 (4%)
Query 102 KDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRC--TDETCVKV 145
+D + G + IGTP Q + I DTGS+NLW+ C E C V
Sbjct 350 EDEAYVGNITIGTPQQQFKVILDTGSSNLWIPDITCGTKPENCSTV 395
> 7300253
Length=465
Score = 40.4 bits (93), Expect = 0.001, Method: Composition-based stats.
Identities = 17/48 (35%), Positives = 27/48 (56%), Gaps = 0/48 (0%)
Query 105 QFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKVTRFDPSK 152
++ + IGTP Q + DTGS+N+WV C + C K ++ P+K
Sbjct 149 EYTCKMNIGTPKQKFTVLPDTGSSNIWVPGPHCKSKACKKHKQYHPAK 196
> CE21684
Length=435
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 18/52 (34%), Positives = 26/52 (50%), Gaps = 0/52 (0%)
Query 91 LAQTSVPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETC 142
L ++P+ D + + +GTPPQ + TGS N WVV + CT C
Sbjct 50 LNSGALPLADYMDEIYTANITVGTPPQSASVVMGTGSANFWVVGAGCTSPAC 101
> CE09542
Length=389
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 20/61 (32%), Positives = 30/61 (49%), Gaps = 1/61 (1%)
Query 91 LAQTSVPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKVTRFDP 150
LA + D ++ G + IGTP Q + DTGS+NLW+ C C ++FD
Sbjct 56 LANLPQNVNDFGDFEYLGNITIGTPDQGFIVVLDTGSSNLWIPGPTCK-TNCKTKSKFDS 114
Query 151 S 151
+
Sbjct 115 T 115
> 7300255
Length=395
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 19/49 (38%), Positives = 30/49 (61%), Gaps = 2/49 (4%)
Query 105 QFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDET--CVKVTRFDPS 151
++ G + IG+P Q +FDTGS NLWV S+ C+ ++ C R++ S
Sbjct 80 EYAGPISIGSPGQPFNMLFDTGSANLWVPSAECSPKSVACHHHHRYNAS 128
> CE05287
Length=428
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 21/65 (32%), Positives = 32/65 (49%), Gaps = 3/65 (4%)
Query 89 LGLAQTSVPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRC---TDETCVKV 145
L LA +S P++ +D + + +G+P Q D+GS+NLWV C D TC
Sbjct 55 LQLASSSSPVIDYEDMAYMVQISLGSPAQNFVLFIDSGSSNLWVPDITCAGGKDATCGSY 114
Query 146 TRFDP 150
+ P
Sbjct 115 CKSTP 119
> CE11480
Length=474
Score = 37.7 bits (86), Expect = 0.008, Method: Composition-based stats.
Identities = 18/52 (34%), Positives = 25/52 (48%), Gaps = 0/52 (0%)
Query 91 LAQTSVPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETC 142
L+ +V D + + IGTP Q + FDT S+NLWV C + C
Sbjct 138 LSTGNVSFFDHFDEYYTAGVRIGTPAQHFQVAFDTTSSNLWVFGVECRSQNC 189
> At1g01300
Length=485
Score = 36.6 bits (83), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 24/87 (27%), Positives = 40/87 (45%), Gaps = 8/87 (9%)
Query 66 AADAPAAPLSFLQHPAQPAATLELGLAQTSVPILQLKDSQFCGMLGIGTPPQWVRPIFDT 125
AA P ++ P ++++ GL+Q S ++ LG+GTP ++V + DT
Sbjct 110 AAQIPGRNVTHAPRPGGFSSSVVSGLSQGS--------GEYFTRLGVGTPARYVYMVLDT 161
Query 126 GSTNLWVVSSRCTDETCVKVTRFDPSK 152
GS +W+ + C FDP K
Sbjct 162 GSDIVWLQCAPCRRCYSQSDPIFDPRK 188
> 7300254
Length=309
Score = 36.6 bits (83), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 16/50 (32%), Positives = 28/50 (56%), Gaps = 2/50 (4%)
Query 105 QFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRC--TDETCVKVTRFDPSK 152
++ G + +G P Q IFDTGS+N W+ S C ++ C +++ S+
Sbjct 2 EYYGTIAMGNPRQNFTVIFDTGSSNTWLPSVNCPMSNSACQNHRKYNSSR 51
> YDR144c
Length=596
Score = 36.6 bits (83), Expect = 0.021, Method: Composition-based stats.
Identities = 17/34 (50%), Positives = 23/34 (67%), Gaps = 0/34 (0%)
Query 102 KDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSS 135
++S + L IGTPPQ V + DTGS++LWV S
Sbjct 77 QNSFYSVELDIGTPPQKVTVLVDTGSSDLWVTGS 110
> Hs22063919
Length=267
Score = 36.2 bits (82), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 16/34 (47%), Positives = 22/34 (64%), Gaps = 0/34 (0%)
Query 97 PILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNL 130
P+ D ++ G +GIGTP Q +FDTGS+NL
Sbjct 67 PLENYLDMEYFGTIGIGTPAQDFTVVFDTGSSNL 100
> At4g22050
Length=336
Score = 35.8 bits (81), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 16/37 (43%), Positives = 23/37 (62%), Gaps = 0/37 (0%)
Query 96 VPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWV 132
V + +KD + G + IG P Q +FDTGS++LWV
Sbjct 36 VQLKNVKDFLYYGKIQIGNPGQTFTVLFDTGSSSLWV 72
> At3g42550
Length=356
Score = 35.4 bits (80), Expect = 0.046, Method: Compositional matrix adjust.
Identities = 17/39 (43%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 112 IGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKVTRFDP 150
IGTPP+ + + DTGS +WV + C VT FDP
Sbjct 84 IGTPPRELDVVIDTGSDLVWVSCNSCVGCPLHNVTFFDP 122
> CE11478
Length=383
Score = 35.0 bits (79), Expect = 0.057, Method: Compositional matrix adjust.
Identities = 15/40 (37%), Positives = 20/40 (50%), Gaps = 0/40 (0%)
Query 103 DSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETC 142
D + + IGTP Q DT S+NLWV C ++C
Sbjct 59 DEYYTVSVRIGTPAQHFEVALDTTSSNLWVFGVECKSQSC 98
> 7296074
Length=393
Score = 34.7 bits (78), Expect = 0.070, Method: Compositional matrix adjust.
Identities = 18/48 (37%), Positives = 27/48 (56%), Gaps = 4/48 (8%)
Query 97 PILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVK 144
P++ D+ F G++ +G Q FDTGS++ WV SS C C+K
Sbjct 79 PLINSYDTNFFGVVSVG--DQSFTMQFDTGSSDFWVPSSHC--RFCIK 122
> YLR120c
Length=569
Score = 34.3 bits (77), Expect = 0.085, Method: Composition-based stats.
Identities = 14/26 (53%), Positives = 20/26 (76%), Gaps = 0/26 (0%)
Query 110 LGIGTPPQWVRPIFDTGSTNLWVVSS 135
L +GTPPQ V + DTGS++LW++ S
Sbjct 87 LEVGTPPQNVTVLVDTGSSDLWIMGS 112
> At5g43100
Length=586
Score = 33.9 bits (76), Expect = 0.13, Method: Composition-based stats.
Identities = 25/74 (33%), Positives = 33/74 (44%), Gaps = 9/74 (12%)
Query 73 PLSFLQHPAQPAAT--LELGLAQTSVPILQLK-------DSQFCGMLGIGTPPQWVRPIF 123
PLS+ P +P L Q+ +P +K + + L IGTPPQ I
Sbjct 34 PLSYSSLPPRPRVEDFRRRRLHQSQLPNAHMKLYDDLLSNGYYTTRLWIGTPPQEFALIV 93
Query 124 DTGSTNLWVVSSRC 137
DTGST +V S C
Sbjct 94 DTGSTVTYVPCSTC 107
> At1g08210
Length=566
Score = 33.9 bits (76), Expect = 0.14, Method: Composition-based stats.
Identities = 17/44 (38%), Positives = 24/44 (54%), Gaps = 5/44 (11%)
Query 112 IGTPPQWVRPIFDTGSTNLWVVSSRC-----TDETCVKVTRFDP 150
+GTPP+ DTGS LWV + C T E ++++ FDP
Sbjct 138 LGTPPREFNVQIDTGSDVLWVSCTSCNGCPKTSELQIQLSFFDP 181
> At2g28040
Length=389
Score = 33.5 bits (75), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 17/43 (39%), Positives = 21/43 (48%), Gaps = 0/43 (0%)
Query 110 LGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKVTRFDPSK 152
L IGTPP + + DTGS ++W C FDPSK
Sbjct 63 LQIGTPPFEIEAVLDTGSEHIWTQCLPCVHCYNQTAPIFDPSK 105
> At3g61820
Length=483
Score = 33.1 bits (74), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 19/45 (42%), Positives = 24/45 (53%), Gaps = 4/45 (8%)
Query 110 LGIGTPPQWVRPIFDTGSTNLWVVSSRCTDETCVKVTR--FDPSK 152
LG+GTP V + DTGS +W+ S C + C T FDP K
Sbjct 139 LGVGTPATNVYMVLDTGSDVVWLQCSPC--KACYNQTDAIFDPKK 181
> YIL015w
Length=587
Score = 32.7 bits (73), Expect = 0.30, Method: Composition-based stats.
Identities = 14/30 (46%), Positives = 19/30 (63%), Gaps = 0/30 (0%)
Query 106 FCGMLGIGTPPQWVRPIFDTGSTNLWVVSS 135
+ L IGTP Q + +FDTGS + WV+ S
Sbjct 45 YATTLDIGTPSQSLTVLFDTGSADFWVMDS 74
> CE09738
Length=389
Score = 32.3 bits (72), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 20/75 (26%), Positives = 34/75 (45%), Gaps = 1/75 (1%)
Query 75 SFLQHPAQPAATLELGLAQTSVPILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWVVS 134
++L++ + A+ LA ++ G + +GTP Q + DTGS NL +
Sbjct 41 AYLEYRDRMVASRSSNLATVVQSTTDYVYYEYMGNITVGTPDQNFIVVLDTGSANLLIPG 100
Query 135 SRCTDETCVKVTRFD 149
+ CT C K F+
Sbjct 101 TNCT-TYCEKKRLFN 114
> At5g45120
Length=491
Score = 31.6 bits (70), Expect = 0.67, Method: Composition-based stats.
Identities = 16/36 (44%), Positives = 22/36 (61%), Gaps = 1/36 (2%)
Query 97 PILQLKDSQFCGMLGIGTPPQWVRPIFDTGSTNLWV 132
P+ +++D + L IGTPPQ V+ DTGS WV
Sbjct 75 PLREVRDG-YLITLNIGTPPQAVQVYLDTGSDLTWV 109
Lambda K H
0.318 0.130 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1925034518
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40