bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
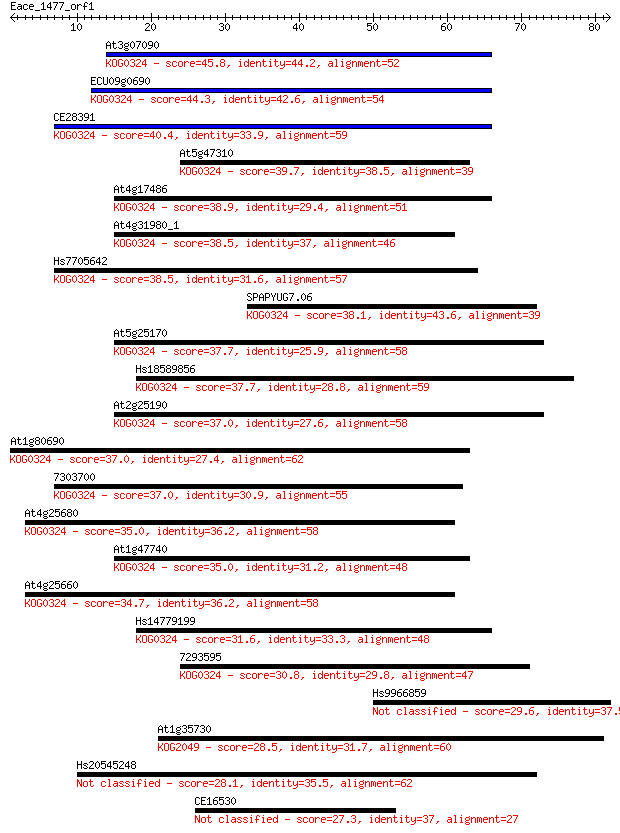
Query= Eace_1477_orf1
Length=81
Score E
Sequences producing significant alignments: (Bits) Value
At3g07090 45.8 2e-05
ECU09g0690 44.3 5e-05
CE28391 40.4 8e-04
At5g47310 39.7 0.001
At4g17486 38.9 0.002
At4g31980_1 38.5 0.003
Hs7705642 38.5 0.003
SPAPYUG7.06 38.1 0.004
At5g25170 37.7 0.005
Hs18589856 37.7 0.006
At2g25190 37.0 0.008
At1g80690 37.0 0.009
7303700 37.0 0.009
At4g25680 35.0 0.033
At1g47740 35.0 0.035
At4g25660 34.7 0.040
Hs14779199 31.6 0.41
7293595 30.8 0.57
Hs9966859 29.6 1.4
At1g35730 28.5 3.4
Hs20545248 28.1 3.9
CE16530 27.3 6.7
> At3g07090
Length=265
Score = 45.8 bits (107), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 23/53 (43%), Positives = 37/53 (69%), Gaps = 3/53 (5%)
Query 14 YGMTPKKIEELGETTKTPQEI-QSFLSSVSPRFTAETYDLLHNNCNHFTQAAA 65
YG TP + ELG + P+++ + +L +SPR+TAE+Y+LL +NCN+F+ A
Sbjct 63 YG-TPIRTIELG-LSHVPKDVFEMYLEEISPRYTAESYNLLTHNCNNFSNEVA 113
> ECU09g0690
Length=150
Score = 44.3 bits (103), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 23/54 (42%), Positives = 30/54 (55%), Gaps = 1/54 (1%)
Query 12 SLYGMTPKKIEELGETTKTPQEIQSFLSSVSPRFTAETYDLLHNNCNHFTQAAA 65
++YG TP KI +LG T + FL S++ F Y LL NNCN+FT A
Sbjct 59 TIYG-TPLKIHDLGATDIPEVVFEDFLFSIAEDFAPHKYHLLKNNCNNFTNTLA 111
> CE28391
Length=334
Score = 40.4 bits (93), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 20/71 (28%), Positives = 33/71 (46%), Gaps = 12/71 (16%)
Query 7 PSEIESLYGMTPKKIEELGETTKTPQ------------EIQSFLSSVSPRFTAETYDLLH 54
P + ++ +P+ EELGET K + +I+ + S+ F + Y L+
Sbjct 71 PYQFSGVFENSPQDAEELGETFKFKESIVVGETERSTSDIRKLIKSLGEDFRGDRYHLIS 130
Query 55 NNCNHFTQAAA 65
NCNHF+ A
Sbjct 131 RNCNHFSAVLA 141
> At5g47310
Length=245
Score = 39.7 bits (91), Expect = 0.001, Method: Composition-based stats.
Identities = 15/39 (38%), Positives = 26/39 (66%), Gaps = 0/39 (0%)
Query 24 LGETTKTPQEIQSFLSSVSPRFTAETYDLLHNNCNHFTQ 62
LG T+ + + +SF+ +S ++ +TY L+ NCNHFT+
Sbjct 94 LGTTSMSRSDFRSFMEKLSRKYHGDTYHLIAKNCNHFTE 132
> At4g17486
Length=246
Score = 38.9 bits (89), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 15/51 (29%), Positives = 29/51 (56%), Gaps = 0/51 (0%)
Query 15 GMTPKKIEELGETTKTPQEIQSFLSSVSPRFTAETYDLLHNNCNHFTQAAA 65
G ++ LG T+ + + +S++ +S ++ +TY L+ NCNHFT+
Sbjct 105 GFIFRRSVLLGTTSMSRSDFRSYMEKLSRKYHGDTYHLIAKNCNHFTEEVC 155
> At4g31980_1
Length=266
Score = 38.5 bits (88), Expect = 0.003, Method: Composition-based stats.
Identities = 17/46 (36%), Positives = 26/46 (56%), Gaps = 0/46 (0%)
Query 15 GMTPKKIEELGETTKTPQEIQSFLSSVSPRFTAETYDLLHNNCNHF 60
G T +K +GET +E++SF+ +S + Y L+ NCNHF
Sbjct 74 GFTFRKSILVGETEMKAKEVRSFMEKLSEEYQGNKYHLITRNCNHF 119
> Hs7705642
Length=193
Score = 38.5 bits (88), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 18/69 (26%), Positives = 30/69 (43%), Gaps = 12/69 (17%)
Query 7 PSEIESLYGMTPKKIEELGETTKTPQ------------EIQSFLSSVSPRFTAETYDLLH 54
P ++ ++P ELGET K + +I+ + + + Y L+H
Sbjct 46 PYPFSGIFEISPGNASELGETFKFKEAVVLGSTDFLEDDIEKIVEELGKEYKGNAYHLMH 105
Query 55 NNCNHFTQA 63
NCNHF+ A
Sbjct 106 KNCNHFSSA 114
> SPAPYUG7.06
Length=191
Score = 38.1 bits (87), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 17/39 (43%), Positives = 21/39 (53%), Gaps = 0/39 (0%)
Query 33 EIQSFLSSVSPRFTAETYDLLHNNCNHFTQAAAAAAAAS 71
++ L +S FT +Y LL NCNHFT AAA S
Sbjct 71 DVDRILIRLSQEFTGLSYSLLERNCNHFTNAAAIELTGS 109
> At5g25170
Length=211
Score = 37.7 bits (86), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 15/58 (25%), Positives = 27/58 (46%), Gaps = 0/58 (0%)
Query 15 GMTPKKIEELGETTKTPQEIQSFLSSVSPRFTAETYDLLHNNCNHFTQAAAAAAAASS 72
G T +K +G T P+ ++ F+ ++ ++ +Y L+ NCNHF S
Sbjct 68 GFTFRKSILIGRTDLDPENVRVFMEKLAEEYSGNSYHLITKNCNHFCNDVCVQLTRRS 125
> Hs18589856
Length=154
Score = 37.7 bits (86), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 17/61 (27%), Positives = 28/61 (45%), Gaps = 2/61 (3%)
Query 18 PKKIEE--LGETTKTPQEIQSFLSSVSPRFTAETYDLLHNNCNHFTQAAAAAAAASSNSN 75
P K +E LG T +I+ + + + Y L+H NCNHF+ A + S+ +
Sbjct 66 PFKFKEVVLGSTDFLEDDIEKIVEELGKEYKGNVYHLMHKNCNHFSSALSEQKLESNEGH 125
Query 76 R 76
Sbjct 126 H 126
> At2g25190
Length=240
Score = 37.0 bits (84), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 16/58 (27%), Positives = 27/58 (46%), Gaps = 0/58 (0%)
Query 15 GMTPKKIEELGETTKTPQEIQSFLSSVSPRFTAETYDLLHNNCNHFTQAAAAAAAASS 72
G T +K +G+T +E++ F+ ++ + Y L+ NCNHF A S
Sbjct 74 GFTFRKSILVGKTDLVAKEVRVFMEKLAEEYQGNKYHLITRNCNHFCNEVCLKLAQKS 131
> At1g80690
Length=231
Score = 37.0 bits (84), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 17/62 (27%), Positives = 32/62 (51%), Gaps = 3/62 (4%)
Query 1 GVVYMPPSEIESLYGMTPKKIEELGETTKTPQEIQSFLSSVSPRFTAETYDLLHNNCNHF 60
G+ P + E G T +K +G+T P E+++ + ++ + +Y+L+ NCNHF
Sbjct 61 GIFEGEPKQCE---GFTFRKSILIGKTDLGPLEVRATMEQLADNYKGSSYNLITKNCNHF 117
Query 61 TQ 62
Sbjct 118 CD 119
> 7303700
Length=205
Score = 37.0 bits (84), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 17/67 (25%), Positives = 34/67 (50%), Gaps = 12/67 (17%)
Query 7 PSEIESLYGMTPKKIEELGE------------TTKTPQEIQSFLSSVSPRFTAETYDLLH 54
P ++ ++P+ +ELG+ T T +E++ + + +F + Y L++
Sbjct 78 PFPFTGVFEISPRDHDELGDQFQFRQSIQIGCTDFTYEEVRRIVEELGNQFRGDRYHLMN 137
Query 55 NNCNHFT 61
NNCNHF+
Sbjct 138 NNCNHFS 144
> At4g25680
Length=252
Score = 35.0 bits (79), Expect = 0.033, Method: Composition-based stats.
Identities = 21/58 (36%), Positives = 28/58 (48%), Gaps = 2/58 (3%)
Query 3 VYMPPSEIESLYGMTPKKIEELGETTKTPQEIQSFLSSVSPRFTAETYDLLHNNCNHF 60
V+ PS +Y K + LG+T T + L +S + TYDLL NCNHF
Sbjct 63 VFSCPSGKNPMYTYREKIV--LGKTDCTIFMVNQMLRELSREWPGHTYDLLSKNCNHF 118
> At1g47740
Length=279
Score = 35.0 bits (79), Expect = 0.035, Method: Composition-based stats.
Identities = 15/48 (31%), Positives = 24/48 (50%), Gaps = 0/48 (0%)
Query 15 GMTPKKIEELGETTKTPQEIQSFLSSVSPRFTAETYDLLHNNCNHFTQ 62
G KK +G T P +++ F+ ++ + Y L+ NCNHF Q
Sbjct 126 GFKFKKSIFIGTTNLNPTQVREFMEDMACSYYGNMYHLIVKNCNHFCQ 173
> At4g25660
Length=255
Score = 34.7 bits (78), Expect = 0.040, Method: Composition-based stats.
Identities = 21/58 (36%), Positives = 28/58 (48%), Gaps = 2/58 (3%)
Query 3 VYMPPSEIESLYGMTPKKIEELGETTKTPQEIQSFLSSVSPRFTAETYDLLHNNCNHF 60
V+ PS +Y K + LG+T T + L +S + TYDLL NCNHF
Sbjct 63 VFSCPSGKNPMYTYREKIV--LGKTDCTIFMVNQILRELSREWPGHTYDLLSKNCNHF 118
> Hs14779199
Length=168
Score = 31.6 bits (70), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 25/49 (51%), Gaps = 1/49 (2%)
Query 18 PKKIEELGETTKTPQEIQSFLSSVSPR-FTAETYDLLHNNCNHFTQAAA 65
P + ++G T T + +LSS+ F E Y+L +NCN F+ A
Sbjct 68 PDSVVDVGSTEVTEEIFLEYLSSLGESLFRGEAYNLFEHNCNTFSNEVA 116
> 7293595
Length=183
Score = 30.8 bits (68), Expect = 0.57, Method: Compositional matrix adjust.
Identities = 14/47 (29%), Positives = 21/47 (44%), Gaps = 0/47 (0%)
Query 24 LGETTKTPQEIQSFLSSVSPRFTAETYDLLHNNCNHFTQAAAAAAAA 70
LG T T E++ ++ + F +Y L NCNHF+ A
Sbjct 98 LGYTDFTCAEVKRVINLLGFEFRGTSYHLTSKNCNHFSNCLAHLVCG 144
> Hs9966859
Length=168
Score = 29.6 bits (65), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 12/32 (37%), Positives = 15/32 (46%), Gaps = 0/32 (0%)
Query 50 YDLLHNNCNHFTQAAAAAAAASSNSNRTTAAV 81
Y+LL NNC HF S +NR + V
Sbjct 112 YNLLVNNCEHFVTLLRYGEGVSEQANRAISTV 143
> At1g35730
Length=564
Score = 28.5 bits (62), Expect = 3.4, Method: Composition-based stats.
Identities = 19/60 (31%), Positives = 27/60 (45%), Gaps = 3/60 (5%)
Query 21 IEELGETTKTPQEIQSFLSSVSPRFTAETYDLLHNNCNHFTQAAAAAAAASSNSNRTTAA 80
++++ ET KT Q+I S + P F A DL N NH Q+ + N AA
Sbjct 342 VQKMIETVKTKQQIALVKSGLKPGFLALVKDL---NGNHVIQSCLQTLGPNDNEFVLEAA 398
> Hs20545248
Length=120
Score = 28.1 bits (61), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 28/66 (42%), Gaps = 5/66 (7%)
Query 10 IESLYGMTPKKIEELGETTKTPQEIQSFLSSVSPRFTAETYDLLHNN----CNHFTQAAA 65
+E L G TPK + LGE KT Q+ + P F + + H NH T A
Sbjct 10 LEPLQG-TPKWVPVLGELQKTLQKGEYLPLRPLPMFESNFVQVTHQGGPVFVNHRTNRLA 68
Query 66 AAAAAS 71
AAS
Sbjct 69 MGVAAS 74
> CE16530
Length=307
Score = 27.3 bits (59), Expect = 6.7, Method: Composition-based stats.
Identities = 10/27 (37%), Positives = 20/27 (74%), Gaps = 0/27 (0%)
Query 26 ETTKTPQEIQSFLSSVSPRFTAETYDL 52
+ T++ +++SF ++++PRFTA T L
Sbjct 84 DRTRSTVDVRSFTTTINPRFTAATKRL 110
Lambda K H
0.308 0.120 0.330
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1162440328
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40