bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1480_orf1
Length=95
Score E
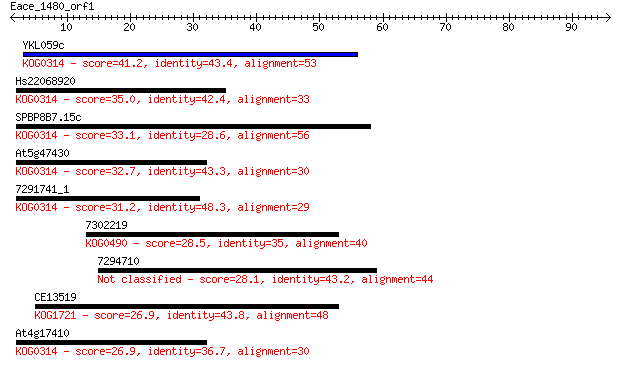
Sequences producing significant alignments: (Bits) Value
YKL059c 41.2 4e-04
Hs22068920 35.0 0.033
SPBP8B7.15c 33.1 0.12
At5g47430 32.7 0.16
7291741_1 31.2 0.48
7302219 28.5 3.2
7294710 28.1 4.0
CE13519 26.9 7.9
At4g17410 26.9 9.3
> YKL059c
Length=441
Score = 41.2 bits (95), Expect = 4e-04, Method: Composition-based stats.
Identities = 23/61 (37%), Positives = 34/61 (55%), Gaps = 9/61 (14%)
Query 3 HHIKNCPKSSDAK-QQKKIRPATGLPSSFLRDI-------LPSEIHKYQEVYIKKDGSFA 54
H IKNCP +SD + K+IR TG+P FL+ I P E+ + +++ I +G F
Sbjct 190 HWIKNCPTNSDPNFEGKRIRRTTGIPKKFLKSIEIDPETMTPEEMAQ-RKIMITDEGKFV 248
Query 55 V 55
V
Sbjct 249 V 249
> Hs22068920
Length=1663
Score = 35.0 bits (79), Expect = 0.033, Method: Composition-based stats.
Identities = 14/35 (40%), Positives = 24/35 (68%), Gaps = 2/35 (5%)
Query 2 GHHIKNCPKSSDAKQQK--KIRPATGLPSSFLRDI 34
GH+IKNCP + D + +I+ +TG+P SF+ ++
Sbjct 73 GHYIKNCPTNGDKNFESGPRIKKSTGIPRSFMMEV 107
> SPBP8B7.15c
Length=482
Score = 33.1 bits (74), Expect = 0.12, Method: Composition-based stats.
Identities = 16/58 (27%), Positives = 30/58 (51%), Gaps = 2/58 (3%)
Query 2 GHHIKNCPKSSDAKQQKK--IRPATGLPSSFLRDILPSEIHKYQEVYIKKDGSFAVLK 57
GH I+ CP ++D K ++ TG+P SFL+++ + I +G + V++
Sbjct 192 GHWIQACPTNADPNYDGKPRVKRTTGIPRSFLKNVERPAEGDAANIMINAEGDYVVVQ 249
> At5g47430
Length=879
Score = 32.7 bits (73), Expect = 0.16, Method: Composition-based stats.
Identities = 13/31 (41%), Positives = 20/31 (64%), Gaps = 1/31 (3%)
Query 2 GHHIKNCPKSSDAKQQ-KKIRPATGLPSSFL 31
GH I++CP + D K+++P TG+P S L
Sbjct 217 GHFIQHCPTNGDPNYDVKRVKPPTGIPKSML 247
> 7291741_1
Length=286
Score = 31.2 bits (69), Expect = 0.48, Method: Composition-based stats.
Identities = 14/29 (48%), Positives = 19/29 (65%), Gaps = 1/29 (3%)
Query 2 GHHIKNCPKSSDAKQQKKIRPATGLPSSF 30
GH IKNCP K Q++++ TG+P SF
Sbjct 160 GHWIKNCPFVG-GKDQQEVKRNTGIPRSF 187
> 7302219
Length=469
Score = 28.5 bits (62), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 23/40 (57%), Gaps = 0/40 (0%)
Query 13 DAKQQKKIRPATGLPSSFLRDILPSEIHKYQEVYIKKDGS 52
DAK K++ TGLP L+ + K++ + +K+DGS
Sbjct 393 DAKDLKQLSQKTGLPKRVLQVWFQNARAKWRRMMMKQDGS 432
> 7294710
Length=537
Score = 28.1 bits (61), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 19/54 (35%), Positives = 27/54 (50%), Gaps = 10/54 (18%)
Query 15 KQQKKIRPATGLPSSFLRDILPSEIHK----------YQEVYIKKDGSFAVLKT 58
K+ +K + PSS +RD+L SE+ K Y+E+Y KK V KT
Sbjct 56 KKSQKFPAGSVQPSSSIRDLLTSELQKSRFMSFKEKFYEEMYFKKATLGGVKKT 109
> CE13519
Length=351
Score = 26.9 bits (58), Expect = 7.9, Method: Composition-based stats.
Identities = 21/48 (43%), Positives = 27/48 (56%), Gaps = 4/48 (8%)
Query 5 IKNCPKSSDAKQQKKIRPATGLPSSFLRDILPSEIHKYQEVYIKKDGS 52
IKN +SDAK K RP + S L P EI KY+EV +K++ S
Sbjct 114 IKN--GTSDAKSHLK-RPFVPI-SPILPHRAPQEIQKYEEVSVKEEPS 157
> At4g17410
Length=744
Score = 26.9 bits (58), Expect = 9.3, Method: Composition-based stats.
Identities = 11/31 (35%), Positives = 19/31 (61%), Gaps = 1/31 (3%)
Query 2 GHHIKNCPKSSDAK-QQKKIRPATGLPSSFL 31
GH I++C + + K+++P TG+P S L
Sbjct 128 GHFIQHCSTNGNPNFDVKRVKPPTGIPKSML 158
Lambda K H
0.321 0.136 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1161385214
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40