bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1498_orf1
Length=177
Score E
Sequences producing significant alignments: (Bits) Value
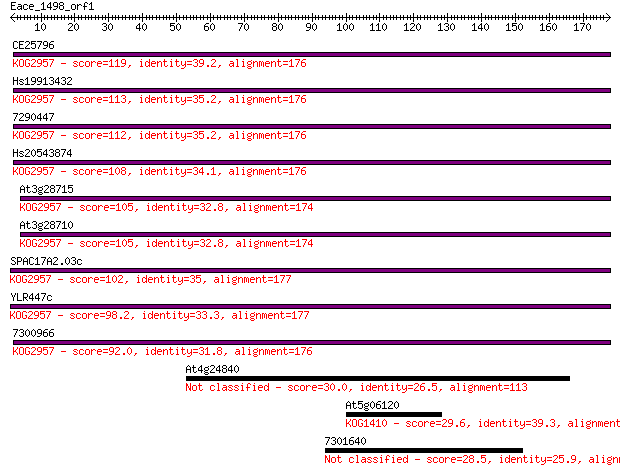
CE25796 119 4e-27
Hs19913432 113 2e-25
7290447 112 5e-25
Hs20543874 108 5e-24
At3g28715 105 4e-23
At3g28710 105 5e-23
SPAC17A2.03c 102 4e-22
YLR447c 98.2 8e-21
7300966 92.0 6e-19
At4g24840 30.0 2.7
At5g06120 29.6 3.5
7301640 28.5 6.6
> CE25796
Length=343
Score = 119 bits (297), Expect = 4e-27, Method: Compositional matrix adjust.
Identities = 69/176 (39%), Positives = 99/176 (56%), Gaps = 23/176 (13%)
Query 2 RSKLTAEFRYLQSQAYQPLAKFLDYIAADKMIDNMLGLIQGALNRKPAEELLARADPLGY 61
+ KL EF +L++ A +PLA FLDYI MIDN++ LI G L+++P EL+ + PLG
Sbjct 67 KEKLVTEFTHLRNNALEPLATFLDYITYSYMIDNIILLITGTLHQRPISELINKCHPLGS 126
Query 62 FGEMRAIAQMDFSSSYEELYRSLLIDTPVGRYFDIFLNNTAAAATPDNPTGHSLRQIEHI 121
F +M AI +S+ ELY ++L+DTP+ YF +N E
Sbjct 127 FEQMEAI---HIASTPAELYNAVLVDTPLANYFVDCIN-------------------EQD 164
Query 122 MTEADVDLLRNSLKKGWLEDFYGFVQTLEDSTTKEVMTHILKTEADFRVLSLAINS 177
+ E +V+++RN+L K ++EDFY F L T EVM IL EAD R + + INS
Sbjct 165 LDEMNVEVIRNTLYKAYIEDFYKFCAGL-GGKTAEVMCDILAFEADRRSIIITINS 219
> Hs19913432
Length=351
Score = 113 bits (282), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 62/176 (35%), Positives = 100/176 (56%), Gaps = 23/176 (13%)
Query 2 RSKLTAEFRYLQSQAYQPLAKFLDYIAADKMIDNMLGLIQGALNRKPAEELLARADPLGY 61
+ K+ EFR++++ AY+PLA FLD+I MIDN++ LI G L+++ EL+ + PLG
Sbjct 75 KEKMVVEFRHMRNHAYEPLASFLDFITYSYMIDNVILLITGTLHQRSIAELVPKCHPLGS 134
Query 62 FGEMRAIAQMDFSSSYEELYRSLLIDTPVGRYFDIFLNNTAAAATPDNPTGHSLRQIEHI 121
F +M A+ + + + ELY ++L+DTP+ +F ++ E
Sbjct 135 FEQMEAV---NIAQTPAELYNAILVDTPLAAFFQDCIS-------------------EQD 172
Query 122 MTEADVDLLRNSLKKGWLEDFYGFVQTLEDSTTKEVMTHILKTEADFRVLSLAINS 177
+ E +++++RN+L K +LE FY F TL TT + M IL+ EAD R + INS
Sbjct 173 LDEMNIEIIRNTLYKAYLESFYKFC-TLLGGTTADAMCPILEFEADRRAFIITINS 227
> 7290447
Length=350
Score = 112 bits (279), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 62/176 (35%), Positives = 100/176 (56%), Gaps = 23/176 (13%)
Query 2 RSKLTAEFRYLQSQAYQPLAKFLDYIAADKMIDNMLGLIQGALNRKPAEELLARADPLGY 61
R KL EF+++++ A +PL+ FLD+I MIDN++ LI G L+++P EL+ + PLG
Sbjct 74 REKLVIEFQHMRNHAVEPLSNFLDFITYGYMIDNIILLITGTLHQRPISELIPKCHPLGS 133
Query 62 FGEMRAIAQMDFSSSYEELYRSLLIDTPVGRYFDIFLNNTAAAATPDNPTGHSLRQIEHI 121
F +M AI +S+ ELY ++L+DTP+ +F ++ E
Sbjct 134 FEQMEAI---HVASTPAELYNAVLVDTPLAPFFVDCIS-------------------EQD 171
Query 122 MTEADVDLLRNSLKKGWLEDFYGFVQTLEDSTTKEVMTHILKTEADFRVLSLAINS 177
+ E +++++RN+L K +LE FY F + + T +VM IL EAD R + + INS
Sbjct 172 LDEMNIEIIRNTLYKAYLEAFYNFCKNM-GGATADVMCEILAFEADRRAIIITINS 226
> Hs20543874
Length=350
Score = 108 bits (270), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 60/176 (34%), Positives = 96/176 (54%), Gaps = 23/176 (13%)
Query 2 RSKLTAEFRYLQSQAYQPLAKFLDYIAADKMIDNMLGLIQGALNRKPAEELLARADPLGY 61
R +L EF Y ++ + +PL+ FL Y+ MIDN++ L+ GAL +K +E+L + PLG
Sbjct 75 RKRLCGEFEYFRNHSLEPLSTFLTYMTCSYMIDNVILLMNGALQKKSVKEILGKCHPLGR 134
Query 62 FGEMRAIAQMDFSSSYEELYRSLLIDTPVGRYFDIFLNNTAAAATPDNPTGHSLRQIEHI 121
F EM A+ + + + +L+ ++LI+TP+ +F ++ A
Sbjct 135 FTEMEAV---NIAETPSDLFNAILIETPLAPFFQDCMSENA------------------- 172
Query 122 MTEADVDLLRNSLKKGWLEDFYGFVQTLEDSTTKEVMTHILKTEADFRVLSLAINS 177
+ E +++LLRN L K +LE FY F + D T EVM IL+ EAD R + +NS
Sbjct 173 LDELNIELLRNKLYKSYLEAFYKFCKNHGD-VTAEVMCPILEFEADRRAFIITLNS 227
> At3g28715
Length=351
Score = 105 bits (263), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 57/174 (32%), Positives = 100/174 (57%), Gaps = 23/174 (13%)
Query 4 KLTAEFRYLQSQAYQPLAKFLDYIAADKMIDNMLGLIQGALNRKPAEELLARADPLGYFG 63
KL +++++ QA +P++ FL+YI MIDN++ ++ G L+ + +EL+ + PLG F
Sbjct 77 KLVDDYKHMLCQATEPMSTFLEYIRYGHMIDNVVLIVTGTLHERDVQELIEKCHPLGMFD 136
Query 64 EMRAIAQMDFSSSYEELYRSLLIDTPVGRYFDIFLNNTAAAATPDNPTGHSLRQIEHIMT 123
+IA + + + ELYR +L+DTP+ YF + T ++ +
Sbjct 137 ---SIATLAVAQNMRELYRLVLVDTPLAPYF-------SECLTSED------------LD 174
Query 124 EADVDLLRNSLKKGWLEDFYGFVQTLEDSTTKEVMTHILKTEADFRVLSLAINS 177
+ +++++RN+L K +LEDFY F Q L T E+M+ +L EAD R +++ INS
Sbjct 175 DMNIEIMRNTLYKAYLEDFYNFCQKL-GGATAEIMSDLLAFEADRRAVNITINS 227
> At3g28710
Length=351
Score = 105 bits (262), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 57/174 (32%), Positives = 100/174 (57%), Gaps = 23/174 (13%)
Query 4 KLTAEFRYLQSQAYQPLAKFLDYIAADKMIDNMLGLIQGALNRKPAEELLARADPLGYFG 63
KL +++++ QA +P++ FL+YI MIDN++ ++ G L+ + +EL+ + PLG F
Sbjct 77 KLVDDYKHMLCQATEPMSTFLEYIRYGHMIDNVVLIVTGTLHERDVQELIEKCHPLGMFD 136
Query 64 EMRAIAQMDFSSSYEELYRSLLIDTPVGRYFDIFLNNTAAAATPDNPTGHSLRQIEHIMT 123
+IA + + + ELYR +L+DTP+ YF + T ++ +
Sbjct 137 ---SIATLAVAQNMRELYRLVLVDTPLAPYF-------SECLTSED------------LD 174
Query 124 EADVDLLRNSLKKGWLEDFYGFVQTLEDSTTKEVMTHILKTEADFRVLSLAINS 177
+ +++++RN+L K +LEDFY F Q L T E+M+ +L EAD R +++ INS
Sbjct 175 DMNIEIMRNTLYKAYLEDFYKFCQKL-GGATAEIMSDLLAFEADRRAVNITINS 227
> SPAC17A2.03c
Length=343
Score = 102 bits (254), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 62/177 (35%), Positives = 99/177 (55%), Gaps = 23/177 (12%)
Query 1 ARSKLTAEFRYLQSQAYQPLAKFLDYIAADKMIDNMLGLIQGALNRKPAEELLARADPLG 60
A KL EF ++ QA + L+KF+DYI MIDN++ L+ G +N + +LL R PLG
Sbjct 70 ATEKLLDEFDLIRRQADETLSKFMDYITYAYMIDNIMLLLTGTVNGQDTHDLLERCHPLG 129
Query 61 YFGEMRAIAQMDFSSSYEELYRSLLIDTPVGRYFDIFLNNTAAAATPDNPTGHSLRQIEH 120
+F + A+ +++ EELY +LI+TP+ YF L + D+
Sbjct 130 WFETLPALC---VATNVEELYSVVLIETPLAPYFKDCL-------SADD----------- 168
Query 121 IMTEADVDLLRNSLKKGWLEDFYGFVQTLEDSTTKEVMTHILKTEADFRVLSLAINS 177
+ E ++++RN+L K +LEDFY F + + + T + M IL+ EAD R +++ INS
Sbjct 169 -LDEQHIEIIRNTLYKAYLEDFYNFCKKI-GACTADTMLPILEFEADRRAITITINS 223
> YLR447c
Length=345
Score = 98.2 bits (243), Expect = 8e-21, Method: Compositional matrix adjust.
Identities = 59/177 (33%), Positives = 92/177 (51%), Gaps = 21/177 (11%)
Query 1 ARSKLTAEFRYLQSQAYQPLAKFLDYIAADKMIDNMLGLIQGALNRKPAEELLARADPLG 60
A SKL EF Y++ Q+ KF+DYI MIDN+ +I G ++ + E+L R PLG
Sbjct 72 ASSKLYHEFNYIRDQSSGSTRKFMDYITYGYMIDNVALMITGTIHDRDKGEILQRCHPLG 131
Query 61 YFGEMRAIAQMDFSSSYEELYRSLLIDTPVGRYFDIFLNNTAAAATPDNPTGHSLRQIEH 120
+F + ++ ++ E LY ++L+DTP+ YF N A
Sbjct 132 WFDTLPTLS---VATDLESLYETVLVDTPLAPYF----KNCFDTAEE------------- 171
Query 121 IMTEADVDLLRNSLKKGWLEDFYGFVQTLEDSTTKEVMTHILKTEADFRVLSLAINS 177
+ + +++++RN L K +LEDFY FV KE M +L EAD R +++A+NS
Sbjct 172 -LDDMNIEIIRNKLYKAYLEDFYNFVTEEIPEPAKECMQTLLGFEADRRSINIALNS 227
> 7300966
Length=350
Score = 92.0 bits (227), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 56/176 (31%), Positives = 95/176 (53%), Gaps = 23/176 (13%)
Query 2 RSKLTAEFRYLQSQAYQPLAKFLDYIAADKMIDNMLGLIQGALNRKPAEELLARADPLGY 61
R +L ++ Y++S + +PL F+++I MIDN+ L+ G N + + LL PLG
Sbjct 71 RDRLLQQYYYIRSHSTEPLTTFMEFIRYPFMIDNVALLVAGLNNHRSMKRLLRMCHPLGE 130
Query 62 FGEMRAIAQMDFSSSYEELYRSLLIDTPVGRYFDIFLNNTAAAATPDNPTGHSLRQIEHI 121
F ++ AI + +S+ EL+ ++LIDTP+ R+ P + SLR +
Sbjct 131 FDQLGAI---EVASNSAELFDAVLIDTPIARF------------VPRDLPMESLRYL--- 172
Query 122 MTEADVDLLRNSLKKGWLEDFYGFVQTLEDSTTKEVMTHILKTEADFRVLSLAINS 177
DV+++R L + +LE FY + L T VMT++L EAD R +++A+N+
Sbjct 173 ----DVEIVRAHLYRAYLEKFYAYCSQL-GGNTANVMTNLLSFEADRRTITIAVNA 223
> At4g24840
Length=724
Score = 30.0 bits (66), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 30/114 (26%), Positives = 54/114 (47%), Gaps = 6/114 (5%)
Query 53 LARADPLGYFGEMRAIAQMDFSSSYEELYRSLLIDTPVGRYFDIFLNNTAAAATPDNPTG 112
L +D + +RA A +D +++ EE++R+ ++ P + T AA T ++
Sbjct 256 LNNSDTSVLYNCLRAYAAIDNTNAAEEIFRTTIV-APFIQKIITHETTTNAAGTSEDELE 314
Query 113 HSLRQIEHIMTEADVDLLR-NSLKKGWLEDFYGFVQTLEDSTTKEVMTHILKTE 165
+ +QI+H + + LL +S K L F L +S KEV+ I K +
Sbjct 315 NDYKQIKHFIAKDCKMLLEISSTDKSGLHVF----DFLANSILKEVLWAIQKVK 364
> At5g06120
Length=1059
Score = 29.6 bits (65), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 11/28 (39%), Positives = 17/28 (60%), Gaps = 0/28 (0%)
Query 100 NTAAAATPDNPTGHSLRQIEHIMTEADV 127
N+ A+ PD+PT H L ++E + E D
Sbjct 405 NSVQASVPDDPTDHPLDKVEVLQDELDC 432
> 7301640
Length=393
Score = 28.5 bits (62), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 15/60 (25%), Positives = 29/60 (48%), Gaps = 2/60 (3%)
Query 94 FDIFLNNTAAAATPDNPTGHSL--RQIEHIMTEADVDLLRNSLKKGWLEDFYGFVQTLED 151
++ L+ P P H++ R EH+ + DLLR++ G + FY + + ++D
Sbjct 198 YEFILDRLLVQYEPHEPEFHNISARVFEHLNESKEFDLLRSTRHFGPMAFFYAWHRCIDD 257
Lambda K H
0.320 0.134 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2743263016
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40