bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
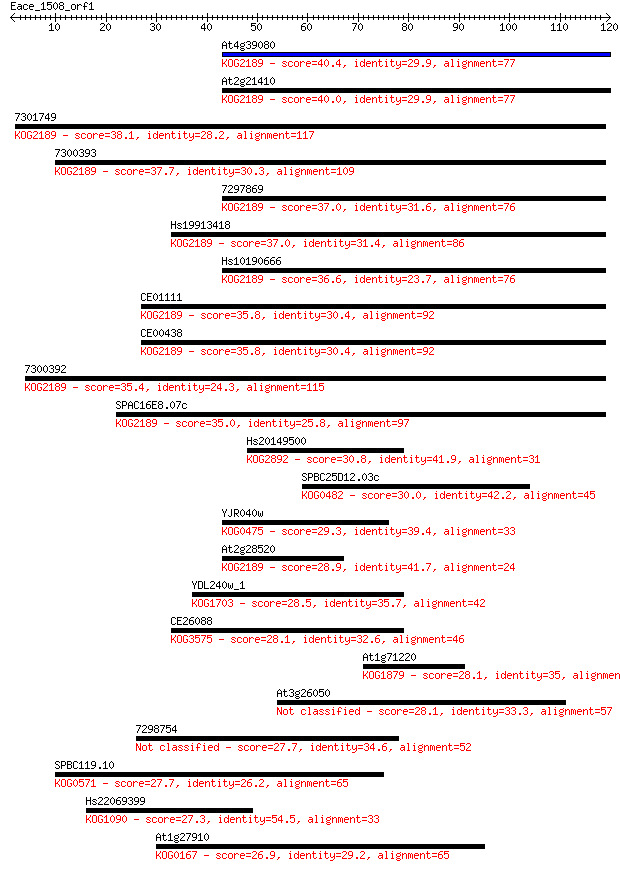
Query= Eace_1508_orf1
Length=119
Score E
Sequences producing significant alignments: (Bits) Value
At4g39080 40.4 8e-04
At2g21410 40.0 0.001
7301749 38.1 0.004
7300393 37.7 0.005
7297869 37.0 0.008
Hs19913418 37.0 0.008
Hs10190666 36.6 0.011
CE01111 35.8 0.020
CE00438 35.8 0.021
7300392 35.4 0.022
SPAC16E8.07c 35.0 0.031
Hs20149500 30.8 0.71
SPBC25D12.03c 30.0 1.1
YJR040w 29.3 2.0
At2g28520 28.9 2.7
YDL240w_1 28.5 2.8
CE26088 28.1 3.6
At1g71220 28.1 4.1
At3g26050 28.1 4.3
7298754 27.7 4.8
SPBC119.10 27.7 5.1
Hs22069399 27.3 7.0
At1g27910 26.9 8.6
> At4g39080
Length=843
Score = 40.4 bits (93), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 23/77 (29%), Positives = 37/77 (48%), Gaps = 4/77 (5%)
Query 43 VAGIVAAKDQERFARAMFRSTRGNAFTHFKRLEEEELTAYPHGEEPKSLFVTYFQGAATG 102
+ G+V + F R +FR+TRGN F +EE + + K++FV ++ G
Sbjct 194 LTGLVPREKSMVFERILFRATRGNIFIRQTVIEEPVIDPNSGEKAEKNVFVVFYSGERAK 253
Query 103 SAFMEKVKCVCSAMGAR 119
S K+ +C A GA
Sbjct 254 S----KILKICEAFGAN 266
> At2g21410
Length=821
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 23/77 (29%), Positives = 37/77 (48%), Gaps = 4/77 (5%)
Query 43 VAGIVAAKDQERFARAMFRSTRGNAFTHFKRLEEEELTAYPHGEEPKSLFVTYFQGAATG 102
+ G+V + F R +FR+TRGN F +EE + + K++FV ++ G
Sbjct 195 LTGLVPREKSMVFERILFRATRGNIFIRQSVIEESVVDPNSGEKAEKNVFVVFYSGERAK 254
Query 103 SAFMEKVKCVCSAMGAR 119
S K+ +C A GA
Sbjct 255 S----KILKICEAFGAN 267
> 7301749
Length=855
Score = 38.1 bits (87), Expect = 0.004, Method: Composition-based stats.
Identities = 33/118 (27%), Positives = 56/118 (47%), Gaps = 16/118 (13%)
Query 2 ADVQQQEKQQTFPYSASRRLLSAGGLGGLESGEMSSLVFSSVAGIVAAKDQERFARAMFR 61
AD Q +++Q +LL G+ + G+ L F VAG++ + F R ++R
Sbjct 165 ADNQNEDEQA--------QLLGEEGVRASQPGQNLKLGF--VAGVILRERLPAFERMLWR 214
Query 62 STRGNAFTHFKRLEEEELTAYPHGEE-PKSLFVTYFQGAATGSAFMEKVKCVCSAMGA 118
+ RGN F + + E L +G++ KS+F+ +FQ G +VK +C A
Sbjct 215 ACRGNVFLR-QAMIETPLEDPTNGDQVHKSVFIIFFQ----GDQLKTRVKKICEGFRA 267
> 7300393
Length=815
Score = 37.7 bits (86), Expect = 0.005, Method: Composition-based stats.
Identities = 33/111 (29%), Positives = 50/111 (45%), Gaps = 13/111 (11%)
Query 10 QQTFPYSASRRLLSAGGLGGLESGEMSSLVFSSVAGIVAAKDQERFARAMFRSTRGNAFT 69
Q+ +S R AGG + L F VAG++ + F R ++R +RGN F
Sbjct 143 QEVLNLDSSNR---AGGDNDAAAQHRGRLGF--VAGVINRERVFAFERMLWRISRGNVF- 196
Query 70 HFKRLEEEELTAYPHGEEP--KSLFVTYFQGAATGSAFMEKVKCVCSAMGA 118
KR + +E P P K++FV +FQ G ++K VC+ A
Sbjct 197 -LKRSDLDEPLNDPATGHPIYKTVFVAFFQ----GEQLKNRIKKVCTGFHA 242
> 7297869
Length=814
Score = 37.0 bits (84), Expect = 0.008, Method: Composition-based stats.
Identities = 24/78 (30%), Positives = 40/78 (51%), Gaps = 8/78 (10%)
Query 43 VAGIVAAKDQERFARAMFRSTRGNAFTHFKRLEEEELTAYPHGEEP--KSLFVTYFQGAA 100
VAG++ + F R ++R +RGN F +R + + L A P K++FV +FQ
Sbjct 175 VAGVIKLERFFSFERMLWRISRGNIF--LRRADIDGLVADEETGRPVLKTVFVAFFQ--- 229
Query 101 TGSAFMEKVKCVCSAMGA 118
G +++K VC+ A
Sbjct 230 -GEQLKQRIKKVCTGYHA 246
> Hs19913418
Length=831
Score = 37.0 bits (84), Expect = 0.008, Method: Composition-based stats.
Identities = 27/90 (30%), Positives = 41/90 (45%), Gaps = 12/90 (13%)
Query 33 GEMSSLVFSSVAGIVAAKDQERFARAMFRSTRGNAFTHFKRLE---EEELTA-YPHGEEP 88
G + L VAG++ + F R ++R RGN F +E E+ +T Y H
Sbjct 161 GRGTPLRLGFVAGVINRERIPTFERMLWRVCRGNVFLRQAEIENPLEDPVTGDYVH---- 216
Query 89 KSLFVTYFQGAATGSAFMEKVKCVCSAMGA 118
KS+F+ +FQ G +VK +C A
Sbjct 217 KSVFIIFFQ----GDQLKNRVKKICEGFRA 242
> Hs10190666
Length=840
Score = 36.6 bits (83), Expect = 0.011, Method: Composition-based stats.
Identities = 18/76 (23%), Positives = 35/76 (46%), Gaps = 4/76 (5%)
Query 43 VAGIVAAKDQERFARAMFRSTRGNAFTHFKRLEEEELTAYPHGEEPKSLFVTYFQGAATG 102
+AG++ + F R ++R RGN + F ++ E K++F+ ++Q G
Sbjct 173 IAGVINRERMASFERLLWRICRGNVYLKFSEMDAPLEDPVTKEEIQKNIFIIFYQ----G 228
Query 103 SAFMEKVKCVCSAMGA 118
+K+K +C A
Sbjct 229 EQLRQKIKKICDGFRA 244
> CE01111
Length=1030
Score = 35.8 bits (81), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 28/97 (28%), Positives = 44/97 (45%), Gaps = 12/97 (12%)
Query 27 LGGLESGEMSSLVFSS--VAGIVAAKDQERFARAMFRSTRGNAFTHFKRLEE---EELTA 81
L G S MS L VAG++ + F R ++R+ RGN F +++ + +T
Sbjct 273 LSGRFSDAMSPLKLQLRFVAGVIQRERLPAFERLLWRACRGNVFLRTSEIDDVLNDTVTG 332
Query 82 YPHGEEPKSLFVTYFQGAATGSAFMEKVKCVCSAMGA 118
P K +F+ +FQG + KVK +C A
Sbjct 333 DPVN---KCVFIIFFQGDHLKT----KVKKICEGFRA 362
> CE00438
Length=935
Score = 35.8 bits (81), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 28/97 (28%), Positives = 44/97 (45%), Gaps = 12/97 (12%)
Query 27 LGGLESGEMSSLVFSS--VAGIVAAKDQERFARAMFRSTRGNAFTHFKRLEE---EELTA 81
L G S MS L VAG++ + F R ++R+ RGN F +++ + +T
Sbjct 178 LSGRFSDAMSPLKLQLRFVAGVIQRERLPAFERLLWRACRGNVFLRTSEIDDVLNDTVTG 237
Query 82 YPHGEEPKSLFVTYFQGAATGSAFMEKVKCVCSAMGA 118
P K +F+ +FQG + KVK +C A
Sbjct 238 DPVN---KCVFIIFFQGDHLKT----KVKKICEGFRA 267
> 7300392
Length=844
Score = 35.4 bits (80), Expect = 0.022, Method: Composition-based stats.
Identities = 28/117 (23%), Positives = 49/117 (41%), Gaps = 8/117 (6%)
Query 4 VQQQEKQQTFPYSASRRLLSAGGLGGLESGEMSSLVFSSVAGIVAAKDQERFARAMFRST 63
+Q E+ F +G E S+ VAG+++ + + F R ++R +
Sbjct 142 IQVLERTDQFFSDQESHNFDLNKMGTHRDPEKSNGHLGFVAGVISREREYAFERMLWRIS 201
Query 64 RGNAFTHFKRLEEEELTAYPHGEE--PKSLFVTYFQGAATGSAFMEKVKCVCSAMGA 118
RGN F +R + + P KS+FV +FQ G +++ VC+ A
Sbjct 202 RGNVFV--RRCDVDVALTDPKTGNVLHKSVFVVFFQ----GDQLQARIRKVCTGFHA 252
> SPAC16E8.07c
Length=805
Score = 35.0 bits (79), Expect = 0.031, Method: Composition-based stats.
Identities = 25/100 (25%), Positives = 49/100 (49%), Gaps = 8/100 (8%)
Query 22 LSAGGLGGLESGEMSSLVFSS---VAGIVAAKDQERFARAMFRSTRGNAFTHFKRLEEEE 78
L G G +S E S + ++ V+GI+ + R ++R+ RGN F H R ++
Sbjct 152 LELGTTGTFDSEETSPQMNTTLDFVSGIIPTVKFQFLERILWRTLRGNLFIHQVRADDSL 211
Query 79 LTAYPHGEEPKSLFVTYFQGAATGSAFMEKVKCVCSAMGA 118
+ EE K++F+ A G+ + +++ + ++GA
Sbjct 212 IHGAEKNEE-KTIFLV----IAHGTQILLRIRKISESLGA 246
> Hs20149500
Length=361
Score = 30.8 bits (68), Expect = 0.71, Method: Composition-based stats.
Identities = 13/31 (41%), Positives = 20/31 (64%), Gaps = 0/31 (0%)
Query 48 AAKDQERFARAMFRSTRGNAFTHFKRLEEEE 78
AA+ Q +F FRS RGN T ++L+E++
Sbjct 151 AAQLQRKFPHLEFRSIRGNLNTRLRKLDEQQ 181
> SPBC25D12.03c
Length=760
Score = 30.0 bits (66), Expect = 1.1, Method: Composition-based stats.
Identities = 19/46 (41%), Positives = 27/46 (58%), Gaps = 2/46 (4%)
Query 59 MFRSTRGNAFTHFKRLEEEELT-AYPHGEEPKSLFVTYFQGAATGS 103
+F STR + F F+ ++ +ELT P G P+SL V + GA T S
Sbjct 241 LFMSTRASKFLPFQEVKIQELTNQVPIGHIPRSLTV-HLYGAITRS 285
> YJR040w
Length=779
Score = 29.3 bits (64), Expect = 2.0, Method: Composition-based stats.
Identities = 13/33 (39%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 43 VAGIVAAKDQERFARAMFRSTRGNAFTHFKRLE 75
+ G+V AKD RF R +R G FT+ + L+
Sbjct 724 LKGLVTAKDILRFKRIKYREVHGAKFTYNEALD 756
> At2g28520
Length=780
Score = 28.9 bits (63), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 10/24 (41%), Positives = 16/24 (66%), Gaps = 0/24 (0%)
Query 43 VAGIVAAKDQERFARAMFRSTRGN 66
++GI+ +F R +FR+TRGN
Sbjct 182 ISGIINKDKLLKFERMLFRATRGN 205
> YDL240w_1
Length=746
Score = 28.5 bits (62), Expect = 2.8, Method: Composition-based stats.
Identities = 15/42 (35%), Positives = 22/42 (52%), Gaps = 8/42 (19%)
Query 37 SLVFSSVAGIVAAKDQERFARAMFRSTRGNAFTHFKRLEEEE 78
+L ++ IVAA+ R R NAF HFK+L+E +
Sbjct 609 TLTLDDISRIVAAEQA--------RELRPNAFAHFKKLKETD 642
> CE26088
Length=511
Score = 28.1 bits (61), Expect = 3.6, Method: Composition-based stats.
Identities = 15/52 (28%), Positives = 27/52 (51%), Gaps = 6/52 (11%)
Query 33 GEMSSLVFSSVAGIVAAKDQERFARAMFR------STRGNAFTHFKRLEEEE 78
G++SS+ + + G + + + R +M + +T GN F F+RL EE
Sbjct 238 GDLSSMTYLKLNGTLHRRARARDVNSMLKLGLIDAATWGNQFDLFRRLSTEE 289
> At1g71220
Length=1674
Score = 28.1 bits (61), Expect = 4.1, Method: Composition-based stats.
Identities = 7/20 (35%), Positives = 17/20 (85%), Gaps = 0/20 (0%)
Query 71 FKRLEEEELTAYPHGEEPKS 90
+++L +E L+++PHG++P +
Sbjct 121 YRQLADESLSSFPHGDDPSA 140
> At3g26050
Length=533
Score = 28.1 bits (61), Expect = 4.3, Method: Composition-based stats.
Identities = 19/58 (32%), Positives = 27/58 (46%), Gaps = 1/58 (1%)
Query 54 RFARAMFRSTRGNAFTHFKRLEEEELTAYPHG-EEPKSLFVTYFQGAATGSAFMEKVK 110
R+A R +AFT + LEE E P E K+ F +F+ A+G A + K
Sbjct 39 RYANETLAWARWSAFTQNRYLEEVERFTKPGSVAEKKAFFEAHFKNRASGKAATQTKK 96
> 7298754
Length=370
Score = 27.7 bits (60), Expect = 4.8, Method: Composition-based stats.
Identities = 18/52 (34%), Positives = 25/52 (48%), Gaps = 6/52 (11%)
Query 26 GLGGLESGEMSSLVFSSVAGIVAAKDQERFARAMFRSTRGNAFTHFKRLEEE 77
G+G ++ G SLV G AKDQE + A R+ H K LE++
Sbjct 121 GIGLIDKGRHCSLVRWRGGGFNNAKDQENYDLARSRT------NHLKMLEDD 166
> SPBC119.10
Length=557
Score = 27.7 bits (60), Expect = 5.1, Method: Composition-based stats.
Identities = 17/65 (26%), Positives = 33/65 (50%), Gaps = 4/65 (6%)
Query 10 QQTFPYSASRRLLSAGGLGGLESGEMSSLVFSSVAGIVAAKDQERFARAMFRSTRGNAFT 69
++T S +RL++ G L SG + S + +S +AA++ E+ A + +S T
Sbjct 215 RETLEASVRKRLMAEVPYGVLLSGGLDSSLIAS----IAARETEKLANSTSQSEEARTIT 270
Query 70 HFKRL 74
+ +L
Sbjct 271 AWPKL 275
> Hs22069399
Length=1531
Score = 27.3 bits (59), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 18/33 (54%), Positives = 21/33 (63%), Gaps = 1/33 (3%)
Query 16 SASRRLLSAGGLGGLESGEMSSLVFSSVAGIVA 48
S SR L GLG LES +SLV +S+AG VA
Sbjct 438 SKSRLLRERAGLGDLESAS-NSLVTNSMAGSVA 469
> At1g27910
Length=768
Score = 26.9 bits (58), Expect = 8.6, Method: Composition-based stats.
Identities = 19/65 (29%), Positives = 29/65 (44%), Gaps = 0/65 (0%)
Query 30 LESGEMSSLVFSSVAGIVAAKDQERFARAMFRSTRGNAFTHFKRLEEEELTAYPHGEEPK 89
L+ G + SLV SV G +++ + +FR R TH + E+T+ G
Sbjct 672 LQEGVIPSLVSISVNGTQRGRERAQKLLTLFRELRQRDQTHLTEPQHTEVTSPEDGFSVA 731
Query 90 SLFVT 94
S VT
Sbjct 732 SAAVT 736
Lambda K H
0.317 0.129 0.359
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1164469306
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40