bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1540_orf1
Length=118
Score E
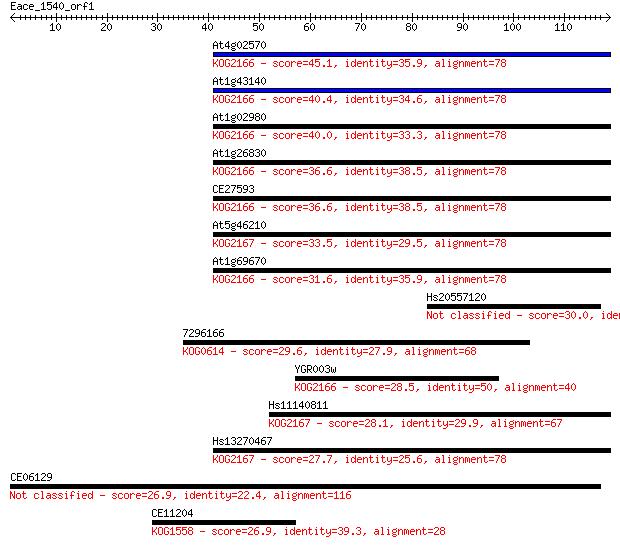
Sequences producing significant alignments: (Bits) Value
At4g02570 45.1 3e-05
At1g43140 40.4 8e-04
At1g02980 40.0 0.001
At1g26830 36.6 0.011
CE27593 36.6 0.012
At5g46210 33.5 0.096
At1g69670 31.6 0.40
Hs20557120 30.0 1.2
7296166 29.6 1.5
YGR003w 28.5 2.8
Hs11140811 28.1 4.1
Hs13270467 27.7 5.0
CE06129 26.9 8.7
CE11204 26.9 8.9
> At4g02570
Length=676
Score = 45.1 bits (105), Expect = 3e-05, Method: Composition-based stats.
Identities = 28/86 (32%), Positives = 44/86 (51%), Gaps = 8/86 (9%)
Query 41 LSVQQLVEDLRLDELTVKKLLGSLMLGRFKILKK-------GENDMFQVNGSFSCLHRKI 93
LS +++ L L + +LL SL ++KIL K +ND F+ N F+ R+I
Sbjct 527 LSYTEILAQLNLSHEDLVRLLHSLSCAKYKILLKEPNTKTVSQNDAFEFNSKFTDRMRRI 586
Query 94 KLP-TPVQEETQCRARVEEDRSITVD 118
K+P PV E + V++DR +D
Sbjct 587 KIPLPPVDERKKVVEDVDKDRRYAID 612
> At1g43140
Length=711
Score = 40.4 bits (93), Expect = 8e-04, Method: Composition-based stats.
Identities = 27/86 (31%), Positives = 43/86 (50%), Gaps = 8/86 (9%)
Query 41 LSVQQLVEDLRLDELTVKKLLGSLMLGRFKILKK-------GENDMFQVNGSFSCLHRKI 93
LS ++ E L L + +LL SL ++KIL K + D F+ N F+ RKI
Sbjct 562 LSYTEISEQLNLSHEDLVRLLHSLSCLKYKILIKEPMSRTISKTDTFEFNSKFTDKMRKI 621
Query 94 KLP-TPVQEETQCRARVEEDRSITVD 118
++P P+ E + V++DR +D
Sbjct 622 RVPLPPMDERKKVVEDVDKDRRYAID 647
> At1g02980
Length=648
Score = 40.0 bits (92), Expect = 0.001, Method: Composition-based stats.
Identities = 26/86 (30%), Positives = 43/86 (50%), Gaps = 8/86 (9%)
Query 41 LSVQQLVEDLRLDELTVKKLLGSLMLGRFKILKK-------GENDMFQVNGSFSCLHRKI 93
LS +++E L L + +LL SL ++KIL K D F+ N F+ R+I
Sbjct 499 LSYTEILEQLNLGHEDLARLLHSLSCLKYKILIKEPMSRNISNTDTFEFNSKFTDKMRRI 558
Query 94 KLP-TPVQEETQCRARVEEDRSITVD 118
++P P+ E + V++DR +D
Sbjct 559 RVPLPPMDERKKIVEDVDKDRRYAID 584
> At1g26830
Length=732
Score = 36.6 bits (83), Expect = 0.011, Method: Composition-based stats.
Identities = 30/92 (32%), Positives = 48/92 (52%), Gaps = 14/92 (15%)
Query 41 LSVQQLVEDLRLDELTVKKLLGSLMLGRFK-ILKK-------GENDMFQVNGSFSCLHRK 92
LS +++ + + +K+ L SL + K ++KK GE D+F VN F+ K
Sbjct 577 LSYKEIEQATEIPAADLKRCLQSLACVKGKNVIKKEPMSKDIGEEDLFVVNDKFTSKFYK 636
Query 93 IKLPTPV-QEET-----QCRARVEEDRSITVD 118
+K+ T V Q+ET + R RVEEDR ++
Sbjct 637 VKIGTVVAQKETEPEKQETRQRVEEDRKPQIE 668
> CE27593
Length=777
Score = 36.6 bits (83), Expect = 0.012, Method: Composition-based stats.
Identities = 30/94 (31%), Positives = 52/94 (55%), Gaps = 16/94 (17%)
Query 41 LSVQQLVEDLRLDELTVKKLLGSLMLGRF--KIL---KKGEN-----DMFQVNGSFSCLH 90
+S QQL+++L++ E +K+ L SL LG+ +IL KG++ D F VN +F
Sbjct 620 ISCQQLMDELKIPERELKRNLQSLALGKASQRILVRKNKGKDAIDMSDEFAVNDNFQSKL 679
Query 91 RKIKLPT---PVQEET---QCRARVEEDRSITVD 118
++K+ V+ E + R +VE+DR + V+
Sbjct 680 TRVKVQMVTGKVESEPEIRETRQKVEDDRKLEVE 713
> At5g46210
Length=617
Score = 33.5 bits (75), Expect = 0.096, Method: Composition-based stats.
Identities = 23/90 (25%), Positives = 45/90 (50%), Gaps = 12/90 (13%)
Query 41 LSVQQLVEDLRLDELTVKKLLGSLMLGRFKILKKG-------ENDMFQVNGSFSC-LHR- 91
LS + + + +++ +++ L SL G+ ++L+K + D F+ N F+ L+R
Sbjct 466 LSSRDIKDSTSIEDKELRRTLQSLACGKVRVLQKNPKGRDVEDGDEFEFNDEFAAPLYRI 525
Query 92 ---KIKLPTPVQEETQCRARVEEDRSITVD 118
I++ V+E T RV +DR +D
Sbjct 526 KVNAIQMKETVEENTSTTERVFQDRQYQID 555
> At1g69670
Length=732
Score = 31.6 bits (70), Expect = 0.40, Method: Composition-based stats.
Identities = 28/92 (30%), Positives = 46/92 (50%), Gaps = 14/92 (15%)
Query 41 LSVQQLVEDLRLDELTVKKLLGSLMLGRFK-ILKK-------GENDMFQVNGSFSCLHRK 92
LS +++ + + +K+ L S+ + K +L+K E D F VN F+ K
Sbjct 577 LSYKEIEQATEIPTPDLKRCLQSMACVKGKNVLRKEPMSKEIAEEDWFVVNDRFASKFYK 636
Query 93 IKLPTPV-QEET-----QCRARVEEDRSITVD 118
+K+ T V Q+ET + R RVEEDR ++
Sbjct 637 VKIGTVVAQKETEPEKQETRQRVEEDRKPQIE 668
> Hs20557120
Length=898
Score = 30.0 bits (66), Expect = 1.2, Method: Composition-based stats.
Identities = 17/41 (41%), Positives = 24/41 (58%), Gaps = 7/41 (17%)
Query 83 NGSFSCLHRKIKLPT-----PVQEETQCR--ARVEEDRSIT 116
+G FSCL+R I +P+ P QE T+ R A+ DR +T
Sbjct 793 HGHFSCLNRNICIPSSTPLAPAQEPTEEREMAKTRRDRWMT 833
> 7296166
Length=768
Score = 29.6 bits (65), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 19/68 (27%), Positives = 32/68 (47%), Gaps = 1/68 (1%)
Query 35 CAGDPALSVQQLVEDLRLDELTVKKLLGSLMLGRFKILKKGENDMFQVNGSFSCLHRKIK 94
C +P +QQ DL+L +L V LG GR +++K D + + CL ++
Sbjct 437 CRDEPKEQLQQEFPDLKLTDLEVVSTLGIGGFGRVELVKAHHQDRVDI-FALKCLKKRHI 495
Query 95 LPTPVQEE 102
+ T +E
Sbjct 496 VDTKQEEH 503
> YGR003w
Length=744
Score = 28.5 bits (62), Expect = 2.8, Method: Composition-based stats.
Identities = 20/44 (45%), Positives = 26/44 (59%), Gaps = 4/44 (9%)
Query 57 VKKLLGSL-MLGRFKILKKGE---NDMFQVNGSFSCLHRKIKLP 96
VK L+ S+ + R KILKK N F VN FS +RK+K+P
Sbjct 605 VKSLVMSMSTIPRCKILKKSSSSGNMKFSVNYFFSSPNRKVKVP 648
> Hs11140811
Length=659
Score = 28.1 bits (61), Expect = 4.1, Method: Composition-based stats.
Identities = 20/81 (24%), Positives = 38/81 (46%), Gaps = 16/81 (19%)
Query 52 LDELTVKKLLGSLMLGRFKILKKG-------ENDMFQVNGSFSCLHR-------KIKLPT 97
+++ +++ L SL G+ ++L K + D F NG F H+ +I++
Sbjct 519 IEDSELRRTLQSLACGKARVLIKSPKGKEVEDGDKFIFNGEFK--HKLFRIKINQIQMKE 576
Query 98 PVQEETQCRARVEEDRSITVD 118
V+E+ RV +DR +D
Sbjct 577 TVEEQVSTTERVFQDRQYQID 597
> Hs13270467
Length=717
Score = 27.7 bits (60), Expect = 5.0, Method: Composition-based stats.
Identities = 20/92 (21%), Positives = 43/92 (46%), Gaps = 16/92 (17%)
Query 41 LSVQQLVEDLRLDELTVKKLLGSLMLGRFKILKKG-------ENDMFQVNGSFSCLHR-- 91
S++++ + +++ +++ L SL G+ ++L K + D F N F H+
Sbjct 566 FSLEEIKQATGIEDGELRRTLQSLACGKARVLAKNPKGKDIEDGDKFICNDDFK--HKLF 623
Query 92 -----KIKLPTPVQEETQCRARVEEDRSITVD 118
+I++ V+E+ RV +DR +D
Sbjct 624 RIKINQIQMKETVEEQASTTERVFQDRQYQID 655
> CE06129
Length=998
Score = 26.9 bits (58), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 26/116 (22%), Positives = 41/116 (35%), Gaps = 18/116 (15%)
Query 1 VGLSLSLSKNSLAIRSCILSFRSFAGGTSGNSGCCAGDPALSVQQLVEDLRLDELTVKKL 60
+G LS+ N+LA + + S +G GN G AL V + + +D +
Sbjct 563 LGAYLSVLINTLAATTTLKSL-DISGNEMGNFGARILSKALQVNVSLRSVSIDNNHI--- 618
Query 61 LGSLMLGRFKILKKGENDMFQVNGSFSCLHRKIKLPTPVQEETQCRARVEEDRSIT 116
G + + S H P PV + C RVE R++
Sbjct 619 --------------GADGFVDLATSIKMNHTLTHFPYPVHDAFDCMQRVERPRTVA 660
> CE11204
Length=477
Score = 26.9 bits (58), Expect = 8.9, Method: Composition-based stats.
Identities = 11/28 (39%), Positives = 16/28 (57%), Gaps = 0/28 (0%)
Query 29 SGNSGCCAGDPALSVQQLVEDLRLDELT 56
SG GCC D +++ +DL +DE T
Sbjct 149 SGAGGCCMADTVDEIKKKEDDLVVDETT 176
Lambda K H
0.319 0.135 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1167969826
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40