bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1547_orf1
Length=152
Score E
Sequences producing significant alignments: (Bits) Value
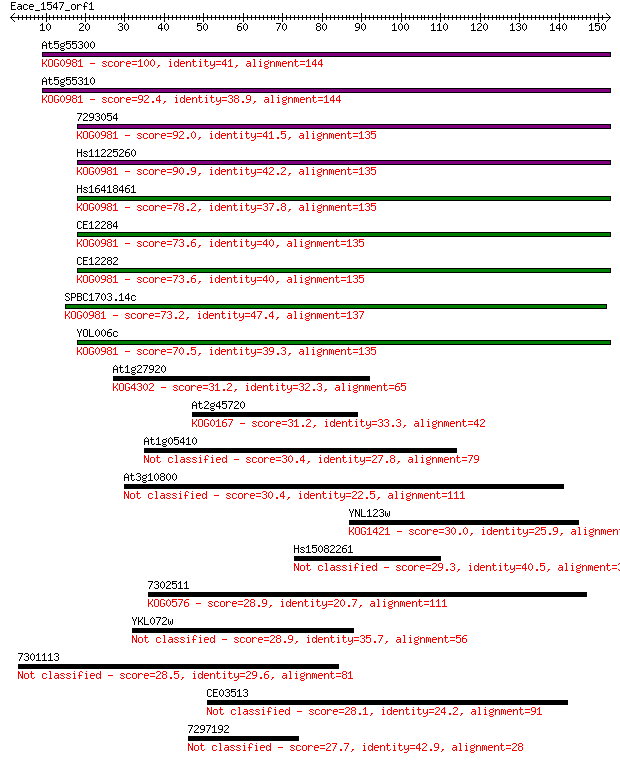
At5g55300 100 8e-22
At5g55310 92.4 3e-19
7293054 92.0 4e-19
Hs11225260 90.9 9e-19
Hs16418461 78.2 6e-15
CE12284 73.6 1e-13
CE12282 73.6 2e-13
SPBC1703.14c 73.2 2e-13
YOL006c 70.5 1e-12
At1g27920 31.2 0.79
At2g45720 31.2 0.87
At1g05410 30.4 1.2
At3g10800 30.4 1.3
YNL123w 30.0 1.7
Hs15082261 29.3 2.8
7302511 28.9 3.7
YKL072w 28.9 4.2
7301113 28.5 5.1
CE03513 28.1 7.5
7297192 27.7 9.9
> At5g55300
Length=916
Score = 100 bits (250), Expect = 8e-22, Method: Compositional matrix adjust.
Identities = 59/144 (40%), Positives = 83/144 (57%), Gaps = 13/144 (9%)
Query 9 SDVNQLVQFYNDANRAVAILCNHQRSIPKQHAASMARLQHQLSAIEEDLRECEEFLAFCK 68
DV Q + Y AN+ VAI+CNHQR++ K H A + +L ++ ++E L+E + L K
Sbjct 755 GDVTQKIVVYQKANKEVAIICNHQRTVSKTHGAQIEKLTARIEELKEVLKELKTNLDRAK 814
Query 69 KDKQQKQFKFKSDFKTLKGEPRKSVVREGMKEDAVLKKIAQLKKKKQTHLLRMHIKDSNK 128
K K L+G K + ++ +A KKIAQ K + MH K+ K
Sbjct 815 KGKPP-----------LEGSDGKKI--RSLEPNAWEKKIAQQSAKIEKMERDMHTKEDLK 861
Query 129 TVALGTSKINYMDPRITVAFCKKY 152
TVALGTSKINY+DPRITVA+CK++
Sbjct 862 TVALGTSKINYLDPRITVAWCKRH 885
> At5g55310
Length=917
Score = 92.4 bits (228), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 56/144 (38%), Positives = 79/144 (54%), Gaps = 13/144 (9%)
Query 9 SDVNQLVQFYNDANRAVAILCNHQRSIPKQHAASMARLQHQLSAIEEDLRECEEFLAFCK 68
DV + V Y AN+ VAI+CNHQR++ K H A + +L ++ + E ++E L K
Sbjct 753 GDVPEKVVVYQQANKEVAIICNHQRTVSKSHGAQVEKLAVKIEELREQIKELNIDLDRAK 812
Query 69 KDKQQKQFKFKSDFKTLKGEPRKSVVREGMKEDAVLKKIAQLKKKKQTHLLRMHIKDSNK 128
K + SD K + + +A+ KKI Q + K + M K+ K
Sbjct 813 KGRTPL---MGSDGKR----------KRNLTPEALEKKIMQTQGKIEKMERDMQTKEDMK 859
Query 129 TVALGTSKINYMDPRITVAFCKKY 152
TVALGTSKINYMDPRITVA+CK++
Sbjct 860 TVALGTSKINYMDPRITVAWCKRH 883
> 7293054
Length=974
Score = 92.0 bits (227), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 56/135 (41%), Positives = 75/135 (55%), Gaps = 32/135 (23%)
Query 18 YNDANRAVAILCNHQRSIPKQHAASMARLQHQLSAIEEDLRECEEFLAFCKKDKQQKQFK 77
YN ANRAVAILCNHQRS+PK H SM L+ ++ A E + +CE + +K+ KQ +
Sbjct 843 YNRANRAVAILCNHQRSVPKSHEKSMENLKEKIKAKREAIEKCESEY-HSRDEKKGKQLE 901
Query 78 FKSDFKTLKGEPRKSVVREGMKEDAVLKKIAQLKKKKQTHLLRMHIKDSNKTVALGTSKI 137
D QLKK + L+ +D NKT+ALGTSK+
Sbjct 902 RLRD---------------------------QLKKLE----LQETDRDENKTIALGTSKL 930
Query 138 NYMDPRITVAFCKKY 152
NY+DPRI+VA+CKK+
Sbjct 931 NYLDPRISVAWCKKH 945
> Hs11225260
Length=765
Score = 90.9 bits (224), Expect = 9e-19, Method: Compositional matrix adjust.
Identities = 57/143 (39%), Positives = 78/143 (54%), Gaps = 33/143 (23%)
Query 18 YNDANRAVAILCNHQRSIPKQHAASMARLQHQLSAIEEDLRECEEFLAFCKKDKQQKQFK 77
YN ANRAVAILCNHQR+ PK SM LQ ++ A +E L + L
Sbjct 619 YNRANRAVAILCNHQRAPPKTFEKSMMNLQTKIDAKKEQLADARRDLKSA---------- 668
Query 78 FKSDFKTLKGEPRKSVVREGMKEDAVLKKIAQLKKKK----QTHLLRMHI----KDSNKT 129
K+D K +K DA KK+ + KKK + L+++ + ++ NK
Sbjct 669 -KADAKVMK--------------DAKTKKVVESKKKAVQRLEEQLMKLEVQATDREENKQ 713
Query 130 VALGTSKINYMDPRITVAFCKKY 152
+ALGTSK+NY+DPRITVA+CKK+
Sbjct 714 IALGTSKLNYLDPRITVAWCKKW 736
> Hs16418461
Length=601
Score = 78.2 bits (191), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 51/139 (36%), Positives = 77/139 (55%), Gaps = 25/139 (17%)
Query 18 YNDANRAVAILCNHQRSIPKQHAASMARLQHQLSAIEEDLRECEEFLAFCKKDKQQKQFK 77
YN ANR VAILCNHQR+ P SM LQ ++ A +E + E + + + + K
Sbjct 455 YNRANRVVAILCNHQRATPSTFEKSMQNLQTKIQAKKEQVAEARA-----ELRRARAEHK 509
Query 78 FKSDFKTLKGEPRKSVVREGMKEDAVLKKIAQLKKKKQTHLLRMHI----KDSNKTVALG 133
+ D K+ +VL+K +L +K Q L ++ + K+ NK VALG
Sbjct 510 AQGDGKS----------------RSVLEKKRRLLEKLQEQLAQLSVQATDKEENKQVALG 553
Query 134 TSKINYMDPRITVAFCKKY 152
TSK+NY+DPRI++A+CK++
Sbjct 554 TSKLNYLDPRISIAWCKRF 572
> CE12284
Length=734
Score = 73.6 bits (179), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 54/138 (39%), Positives = 77/138 (55%), Gaps = 34/138 (24%)
Query 18 YNDANRAVAILCNHQRSIPKQHAASMARLQHQLSAIEEDLRECEEFLAFCK---KDKQQK 74
YN ANR VAILCNHQR++ K SM +L+ ++ +++++E E L + K+K QK
Sbjct 596 YNRANRQVAILCNHQRAVSKGFDESMQKLEQKIKDKKKEVKEAEAALKSARGAEKEKAQK 655
Query 75 QFKFKSDFKTLKGEPRKSVVREGMKEDAVLKKIAQLKKKKQTHLLRMHIKDSNKTVALGT 134
++ + +KE QLKK K + KD NK +ALGT
Sbjct 656 KY-------------------DRLKE--------QLKKLK----ISRTDKDENKQIALGT 684
Query 135 SKINYMDPRITVAFCKKY 152
SK+NY+DPRITVA+CKK+
Sbjct 685 SKLNYIDPRITVAWCKKF 702
> CE12282
Length=806
Score = 73.6 bits (179), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 54/138 (39%), Positives = 77/138 (55%), Gaps = 34/138 (24%)
Query 18 YNDANRAVAILCNHQRSIPKQHAASMARLQHQLSAIEEDLRECEEFLAFCK---KDKQQK 74
YN ANR VAILCNHQR++ K SM +L+ ++ +++++E E L + K+K QK
Sbjct 668 YNRANRQVAILCNHQRAVSKGFDESMQKLEQKIKDKKKEVKEAEAALKSARGAEKEKAQK 727
Query 75 QFKFKSDFKTLKGEPRKSVVREGMKEDAVLKKIAQLKKKKQTHLLRMHIKDSNKTVALGT 134
++ + +KE QLKK K + KD NK +ALGT
Sbjct 728 KY-------------------DRLKE--------QLKKLK----ISRTDKDENKQIALGT 756
Query 135 SKINYMDPRITVAFCKKY 152
SK+NY+DPRITVA+CKK+
Sbjct 757 SKLNYIDPRITVAWCKKF 774
> SPBC1703.14c
Length=814
Score = 73.2 bits (178), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 65/197 (32%), Positives = 89/197 (45%), Gaps = 60/197 (30%)
Query 15 VQFYNDANRAVAILCNHQRSIPKQHAASMAR-------LQHQLSAIEEDLRECEEFLAFC 67
+ FYN ANR VAILCNHQRS+ K H M R LQ+Q + + + E LA
Sbjct 589 ILFYNRANRTVAILCNHQRSVTKNHDVQMERFAERIKALQYQRMRLRKMMLNLEPKLAKS 648
Query 68 KKDKQQKQ------------------------FKFKSDFKTLKGEPRKSVVREGMKE--- 100
K + K+ KF + + L E KSV+ E E
Sbjct 649 KPELLAKEEGITDSWIVKHHETLYELEKEKIKKKFDRENEKLAAEDPKSVLPESELEVRL 708
Query 101 ----------DAVLKK---------IAQLKKK-----KQTHLLRMHI--KDSNKTVALGT 134
DA LK + QL+K+ ++ +++R + KD NKT ALGT
Sbjct 709 KAADELKKALDAELKSKKVDPGRSSMEQLEKRLNKLNERINVMRTQMIDKDENKTTALGT 768
Query 135 SKINYMDPRITVAFCKK 151
SKINY+DPR+T +F K+
Sbjct 769 SKINYIDPRLTYSFSKR 785
> YOL006c
Length=769
Score = 70.5 bits (171), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 53/196 (27%), Positives = 85/196 (43%), Gaps = 61/196 (31%)
Query 18 YNDANRAVAILCNHQRSIPKQHAASMARLQHQLSAIEEDLRECEEFLAFCKKDKQQKQFK 77
YN ANR VAILCNHQR++ K HA ++ + +++ +E C+ + KD +K+ K
Sbjct 545 YNAANRTVAILCNHQRTVTKGHAQTVEKANNRIQELEWQKIRCKRAILQLDKDLLKKEPK 604
Query 78 FKSDFKTLKGEPRKSV----------------VREG----------MKEDAVLKKIAQLK 111
+ + L E ++ VRE + E + + + ++
Sbjct 605 YFEEIDDLTKEDEATIHKRIIDREIEKYQRKFVRENDKRKFEKEELLPESQLKEWLEKVD 664
Query 112 KKKQ-----------------------------------THLLRMHIKDSNKTVALGTSK 136
+KKQ T +++ K+ N V+LGTSK
Sbjct 665 EKKQEFEKELKTGEVELKSSWNSVEKIKAQVEKLEQRIQTSSIQLKDKEENSQVSLGTSK 724
Query 137 INYMDPRITVAFCKKY 152
INY+DPR++V FCKKY
Sbjct 725 INYIDPRLSVVFCKKY 740
> At1g27920
Length=592
Score = 31.2 bits (69), Expect = 0.79, Method: Compositional matrix adjust.
Identities = 21/65 (32%), Positives = 34/65 (52%), Gaps = 11/65 (16%)
Query 27 ILCNHQRSIPKQHAASMARLQHQLSAIEEDLRECEEFLAFCKKDKQQKQFKFKSDFKTLK 86
+LC +RS+P + L+ QL +I LRE +KD++ KQ F+++K
Sbjct 115 LLCLGERSVPGRPEKKGGTLREQLDSIAPALREMR-----LRKDERVKQ------FRSVK 163
Query 87 GEPRK 91
GE +K
Sbjct 164 GEIQK 168
> At2g45720
Length=553
Score = 31.2 bits (69), Expect = 0.87, Method: Composition-based stats.
Identities = 14/42 (33%), Positives = 23/42 (54%), Gaps = 0/42 (0%)
Query 47 QHQLSAIEEDLRECEEFLAFCKKDKQQKQFKFKSDFKTLKGE 88
+ QL A+ E L+E E C +KQ+ + K +SD +L +
Sbjct 74 KEQLQAVLETLKETIELANVCVSEKQEGKLKMQSDLDSLSAK 115
> At1g05410
Length=481
Score = 30.4 bits (67), Expect = 1.2, Method: Composition-based stats.
Identities = 22/85 (25%), Positives = 39/85 (45%), Gaps = 6/85 (7%)
Query 35 IPKQHAASMARLQHQLSAIEEDLRECEEFLAFCKKDKQQKQFKFKSDFKTLKGEPRKSVV 94
IP H A M +L+ ++ + LR+ +EF + K Q + SD + + +
Sbjct 365 IPFNHEAEMHKLEEEIGEVLRALRKAQEFEYQIAEGKLHAQKECLSDLYRQLEKEKSELS 424
Query 95 REGMKEDA------VLKKIAQLKKK 113
R DA VLK++ Q++K+
Sbjct 425 RRVSGTDANSLMTNVLKRLDQIRKE 449
> At3g10800
Length=675
Score = 30.4 bits (67), Expect = 1.3, Method: Composition-based stats.
Identities = 25/111 (22%), Positives = 53/111 (47%), Gaps = 17/111 (15%)
Query 30 NHQRSIPKQHAASMARLQHQLSAIEEDLRECEEFLAFCKKDKQQKQFKFKSDFKTLKGEP 89
NHQ+S S+++ + + +LR C+ + K D + + D +
Sbjct 146 NHQKS-------SVSKRKKENGDSSGELRSCK----YQKSDDKSVATNNEGD----DDDD 190
Query 90 RKSVVREGMKEDAVLKKIAQLKKKKQTHLLRMHIKDSNKTVALGTSKINYM 140
++ ++R+ ++ ++++L+KK+QT L +K N T+A KI Y+
Sbjct 191 KRKLIRQIRNRESA--QLSRLRKKQQTEELERKVKSMNATIAELNGKIAYV 239
> YNL123w
Length=997
Score = 30.0 bits (66), Expect = 1.7, Method: Composition-based stats.
Identities = 15/58 (25%), Positives = 34/58 (58%), Gaps = 2/58 (3%)
Query 87 GEPRKSVVREGMKEDAVLKKIAQLKKKKQTHLLRMHIKDSNKTVALGTSKINYMDPRI 144
G+PR S+V G +VL+ A+++ + ++RM + +N+ + S+++Y + +I
Sbjct 753 GKPRVSIVDAGFGSISVLQ--ARIRGVPEEWIMRMEHESNNRLQFITVSRVSYTEDKI 808
> Hs15082261
Length=365
Score = 29.3 bits (64), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 15/37 (40%), Positives = 22/37 (59%), Gaps = 0/37 (0%)
Query 73 QKQFKFKSDFKTLKGEPRKSVVREGMKEDAVLKKIAQ 109
Q QF+F+ TL+G R S+++ G KE A L I+
Sbjct 94 QFQFRFERWNCTLEGRYRASLLKRGFKETAFLYAISS 130
> 7302511
Length=1218
Score = 28.9 bits (63), Expect = 3.7, Method: Composition-based stats.
Identities = 23/111 (20%), Positives = 44/111 (39%), Gaps = 0/111 (0%)
Query 36 PKQHAASMARLQHQLSAIEEDLRECEEFLAFCKKDKQQKQFKFKSDFKTLKGEPRKSVVR 95
PK+ + LQH E LR +E L + Q + D +++ G P++ +
Sbjct 268 PKKRPTAERLLQHPFVQCEMSLRVAKELLQKYQSPNPQFYYYLDGDEESVAGVPQRIASK 327
Query 96 EGMKEDAVLKKIAQLKKKKQTHLLRMHIKDSNKTVALGTSKINYMDPRITV 146
+ + V + LK T+ S +T+ S + Y+D + +
Sbjct 328 MTSRTNGVPAQNHTLKTGMTTNSTWNERSSSPETLPSDMSLLQYIDEELKL 378
> YKL072w
Length=766
Score = 28.9 bits (63), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 20/56 (35%), Positives = 25/56 (44%), Gaps = 0/56 (0%)
Query 32 QRSIPKQHAASMARLQHQLSAIEEDLRECEEFLAFCKKDKQQKQFKFKSDFKTLKG 87
QR I + LQ +SA+E L E E C K ++ KF S KT KG
Sbjct 59 QRKISTLITSYTGNLQDTISAVEVALPEDPEEWPCCLKKYHEELLKFSSPKKTAKG 114
> 7301113
Length=201
Score = 28.5 bits (62), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 24/84 (28%), Positives = 38/84 (45%), Gaps = 3/84 (3%)
Query 3 VLNVDISDVNQLVQF---YNDANRAVAILCNHQRSIPKQHAASMARLQHQLSAIEEDLRE 59
+L DI ++++L QF Y N + L I H AS A ++ Q + EDLR
Sbjct 30 LLQDDIRNLSKLKQFCMNYTVPNNNRSYLWALVMGILPLHKASTAYVRDQRREMYEDLRR 89
Query 60 CEEFLAFCKKDKQQKQFKFKSDFK 83
L F ++++ F + D K
Sbjct 90 AVTVLRFTDHKQKEQPFMWLIDHK 113
> CE03513
Length=396
Score = 28.1 bits (61), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 22/91 (24%), Positives = 31/91 (34%), Gaps = 2/91 (2%)
Query 51 SAIEEDLRECEEFLAFCKKDKQQKQFKFKSDFKTLKGEPRKSVVREGMKEDAVLKKIAQL 110
A REC +C Q K+ D++ PR + + D +KK
Sbjct 306 GAFPSAFRECRNTCGYCSATMQMTVVKY--DYRLATSTPRCDIKTYFARTDVEMKKRYNP 363
Query 111 KKKKQTHLLRMHIKDSNKTVALGTSKINYMD 141
KK+ L R+ K T NY D
Sbjct 364 AKKEFDPLARLIWKRPKDTDYFVVEDHNYED 394
> 7297192
Length=924
Score = 27.7 bits (60), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 12/28 (42%), Positives = 19/28 (67%), Gaps = 0/28 (0%)
Query 46 LQHQLSAIEEDLRECEEFLAFCKKDKQQ 73
LQ A+EEDLR+C++ L+ + D Q+
Sbjct 55 LQSAKGALEEDLRKCKQKLSLAEGDVQR 82
Lambda K H
0.319 0.131 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1925034518
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40