bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
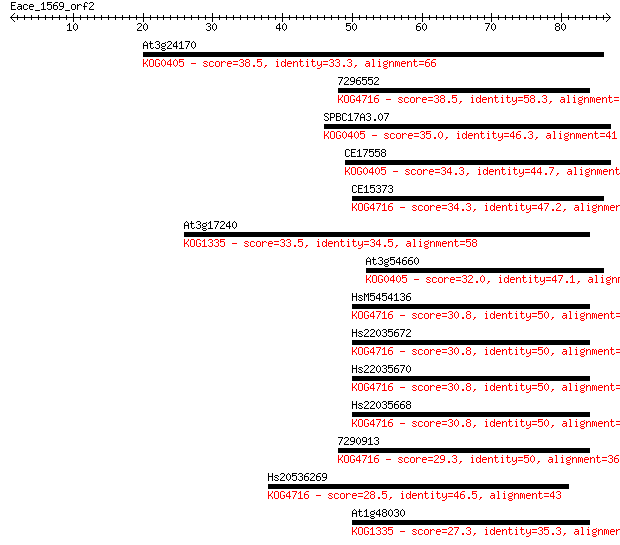
Query= Eace_1569_orf2
Length=86
Score E
Sequences producing significant alignments: (Bits) Value
At3g24170 38.5 0.003
7296552 38.5 0.003
SPBC17A3.07 35.0 0.035
CE17558 34.3 0.059
CE15373 34.3 0.059
At3g17240 33.5 0.11
At3g54660 32.0 0.27
HsM5454136 30.8 0.66
Hs22035672 30.8 0.66
Hs22035670 30.8 0.66
Hs22035668 30.8 0.66
7290913 29.3 2.0
Hs20536269 28.5 2.8
At1g48030 27.3 7.2
> At3g24170
Length=499
Score = 38.5 bits (88), Expect = 0.003, Method: Composition-based stats.
Identities = 22/66 (33%), Positives = 34/66 (51%), Gaps = 6/66 (9%)
Query 20 SSSSSSSSKQQLVVVVQHAENPSITKTIVVDTVLYATGRKADIHNLNLLAVGVKTNEGGK 79
S + + + Q + V+ H E V D VL+ATGR + LNL AVGV+ ++ G
Sbjct 266 SLTQLTKTDQGIKVISSHGEE------FVADVVLFATGRSPNTKRLNLEAVGVELDQAGA 319
Query 80 IICDYF 85
+ D +
Sbjct 320 VKVDEY 325
> 7296552
Length=516
Score = 38.5 bits (88), Expect = 0.003, Method: Composition-based stats.
Identities = 21/36 (58%), Positives = 26/36 (72%), Gaps = 1/36 (2%)
Query 48 VVDTVLYATGRKADIHNLNLLAVGVKTNEGGKIICD 83
V DTVL+A GRK I +LNL A GVKT++ KI+ D
Sbjct 302 VFDTVLWAIGRKGLIEDLNLDAAGVKTHD-DKIVVD 336
> SPBC17A3.07
Length=464
Score = 35.0 bits (79), Expect = 0.035, Method: Composition-based stats.
Identities = 19/41 (46%), Positives = 23/41 (56%), Gaps = 0/41 (0%)
Query 46 TIVVDTVLYATGRKADIHNLNLLAVGVKTNEGGKIICDYFR 86
T VDT+L+A GR I L L GVKT G II D ++
Sbjct 263 TFNVDTLLWAIGRAPKIQGLRLEKAGVKTLPNGIIIADTYQ 303
> CE17558
Length=473
Score = 34.3 bits (77), Expect = 0.059, Method: Compositional matrix adjust.
Identities = 17/38 (44%), Positives = 24/38 (63%), Gaps = 0/38 (0%)
Query 49 VDTVLYATGRKADIHNLNLLAVGVKTNEGGKIICDYFR 86
V T+++A GR LNL VGVKT++ G II D ++
Sbjct 271 VQTLIWAIGRDPLTKELNLERVGVKTDKSGHIIVDEYQ 308
> CE15373
Length=503
Score = 34.3 bits (77), Expect = 0.059, Method: Compositional matrix adjust.
Identities = 17/37 (45%), Positives = 26/37 (70%), Gaps = 1/37 (2%)
Query 50 DTVLYATGRKADIHNLNLLAVGVKTNE-GGKIICDYF 85
DTV++A GR ++ +LNL GV+T++ GKI+ D F
Sbjct 288 DTVIWAAGRVPNLKSLNLDNAGVRTDKRSGKILADEF 324
> At3g17240
Length=507
Score = 33.5 bits (75), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 20/58 (34%), Positives = 34/58 (58%), Gaps = 1/58 (1%)
Query 26 SSKQQLVVVVQHAENPSITKTIVVDTVLYATGRKADIHNLNLLAVGVKTNEGGKIICD 83
SS + ++V+ AE T T+ D VL + GR L+L +GV+T++GG+I+ +
Sbjct 283 SSGDGVKLIVEPAEGGEQT-TLEADVVLVSAGRTPFTSGLDLEKIGVETDKGGRILVN 339
> At3g54660
Length=565
Score = 32.0 bits (71), Expect = 0.27, Method: Composition-based stats.
Identities = 16/34 (47%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 52 VLYATGRKADIHNLNLLAVGVKTNEGGKIICDYF 85
V++ATGRK + NL L VGVK + G I D +
Sbjct 350 VMFATGRKPNTKNLGLENVGVKMAKNGAIEVDEY 383
> HsM5454136
Length=524
Score = 30.8 bits (68), Expect = 0.66, Method: Composition-based stats.
Identities = 17/35 (48%), Positives = 22/35 (62%), Gaps = 1/35 (2%)
Query 50 DTVLYATGRKADIHNLNLLAVGVKTN-EGGKIICD 83
DTVL+A GR D +LNL GV T+ + KI+ D
Sbjct 310 DTVLWAIGRVPDTRSLNLEKAGVDTSPDTQKILVD 344
> Hs22035672
Length=524
Score = 30.8 bits (68), Expect = 0.66, Method: Composition-based stats.
Identities = 17/35 (48%), Positives = 22/35 (62%), Gaps = 1/35 (2%)
Query 50 DTVLYATGRKADIHNLNLLAVGVKTN-EGGKIICD 83
DTVL+A GR D +LNL GV T+ + KI+ D
Sbjct 310 DTVLWAIGRVPDTRSLNLEKAGVDTSPDTQKILVD 344
> Hs22035670
Length=494
Score = 30.8 bits (68), Expect = 0.66, Method: Composition-based stats.
Identities = 17/35 (48%), Positives = 22/35 (62%), Gaps = 1/35 (2%)
Query 50 DTVLYATGRKADIHNLNLLAVGVKTN-EGGKIICD 83
DTVL+A GR D +LNL GV T+ + KI+ D
Sbjct 280 DTVLWAIGRVPDTRSLNLEKAGVDTSPDTQKILVD 314
> Hs22035668
Length=428
Score = 30.8 bits (68), Expect = 0.66, Method: Composition-based stats.
Identities = 17/35 (48%), Positives = 22/35 (62%), Gaps = 1/35 (2%)
Query 50 DTVLYATGRKADIHNLNLLAVGVKTN-EGGKIICD 83
DTVL+A GR D +LNL GV T+ + KI+ D
Sbjct 214 DTVLWAIGRVPDTRSLNLEKAGVDTSPDTQKILVD 248
> 7290913
Length=491
Score = 29.3 bits (64), Expect = 2.0, Method: Composition-based stats.
Identities = 18/36 (50%), Positives = 22/36 (61%), Gaps = 1/36 (2%)
Query 48 VVDTVLYATGRKADIHNLNLLAVGVKTNEGGKIICD 83
V DTVL+A GRK + +LNL GV T + KI D
Sbjct 277 VYDTVLWAIGRKGLVDDLNLPNAGV-TVQKDKIPVD 311
> Hs20536269
Length=459
Score = 28.5 bits (62), Expect = 2.8, Method: Composition-based stats.
Identities = 20/46 (43%), Positives = 27/46 (58%), Gaps = 3/46 (6%)
Query 38 AENPSITKTI--VVDTVLYATGRKADIHNLNLLAVGVKTNE-GGKI 80
A++ T+TI V +TVL A GR + + L +GVK NE GKI
Sbjct 233 AKSTEGTETIEGVYNTVLLAIGRDSCTRKIGLEKIGVKINEKSGKI 278
> At1g48030
Length=507
Score = 27.3 bits (59), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 12/34 (35%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 50 DTVLYATGRKADIHNLNLLAVGVKTNEGGKIICD 83
D VL + GR L+L +GV+T++ G+I+ +
Sbjct 306 DVVLVSAGRTPFTSGLDLEKIGVETDKAGRILVN 339
Lambda K H
0.306 0.121 0.311
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1190857334
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40