bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1572_orf2
Length=126
Score E
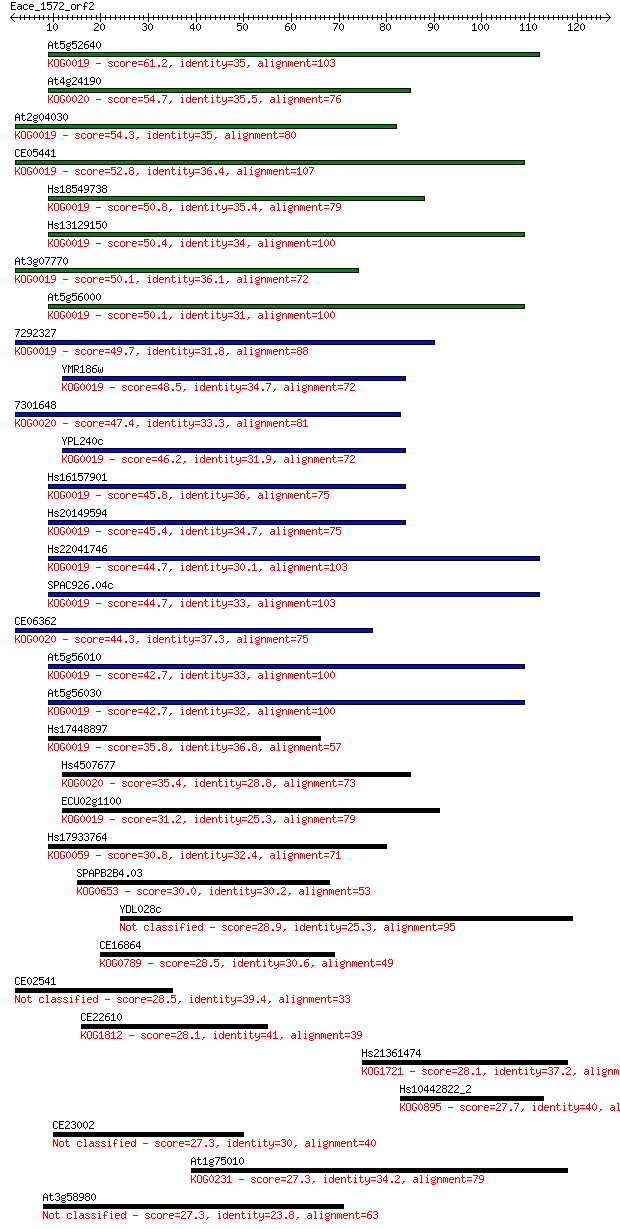
Sequences producing significant alignments: (Bits) Value
At5g52640 61.2 4e-10
At4g24190 54.7 4e-08
At2g04030 54.3 5e-08
CE05441 52.8 2e-07
Hs18549738 50.8 7e-07
Hs13129150 50.4 7e-07
At3g07770 50.1 9e-07
At5g56000 50.1 1e-06
7292327 49.7 1e-06
YMR186w 48.5 3e-06
7301648 47.4 7e-06
YPL240c 46.2 1e-05
Hs16157901 45.8 2e-05
Hs20149594 45.4 3e-05
Hs22041746 44.7 4e-05
SPAC926.04c 44.7 4e-05
CE06362 44.3 6e-05
At5g56010 42.7 2e-04
At5g56030 42.7 2e-04
Hs17448897 35.8 0.021
Hs4507677 35.4 0.023
ECU02g1100 31.2 0.43
Hs17933764 30.8 0.65
SPAPB2B4.03 30.0 1.2
YDL028c 28.9 2.6
CE16864 28.5 2.8
CE02541 28.5 3.3
CE22610 28.1 3.8
Hs21361474 28.1 4.4
Hs10442822_2 27.7 4.7
CE23002 27.3 7.0
At1g75010 27.3 7.0
At3g58980 27.3 7.6
> At5g52640
Length=705
Score = 61.2 bits (147), Expect = 4e-10, Method: Composition-based stats.
Identities = 36/104 (34%), Positives = 60/104 (57%), Gaps = 5/104 (4%)
Query 9 MRGQKVFEINPSHRIMQQLLQRVR-DGGEDSMKDEDTQLAERLFDAAMLASGFDVEQPAQ 67
M +K EINP + IM++L +R D + S+KD L L++ A+L SGF +++P
Sbjct 606 MSSKKTMEINPDNGIMEELRKRAEADKNDKSVKD----LVMLLYETALLTSGFSLDEPNT 661
Query 68 LAAVLYKGLASQVGVDPADPINENIDLPEEEADKTEENAEEPMD 111
AA +++ L + +D + + E+ D+PE E D EE+ E +D
Sbjct 662 FAARIHRMLKLGLSIDEDENVEEDGDMPELEEDAAEESKMEEVD 705
> At4g24190
Length=823
Score = 54.7 bits (130), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 27/76 (35%), Positives = 42/76 (55%), Gaps = 3/76 (3%)
Query 9 MRGQKVFEINPSHRIMQQLLQRVRDGGEDSMKDEDTQLAERLFDAAMLASGFDVEQPAQL 68
MRG++V EINP H I+++L R+ ED E QL ++ A++ SGF + P
Sbjct 702 MRGKRVLEINPRHPIIKELKDRIASDPEDESVKETAQL---MYQTALIESGFILTDPKDF 758
Query 69 AAVLYKGLASQVGVDP 84
AA +Y + S + + P
Sbjct 759 AARIYNSVKSGLNISP 774
> At2g04030
Length=780
Score = 54.3 bits (129), Expect = 5e-08, Method: Composition-based stats.
Identities = 28/80 (35%), Positives = 48/80 (60%), Gaps = 3/80 (3%)
Query 2 DPMHLRLMRGQKVFEINPSHRIMQQLLQRVRDGGEDSMKDEDTQLAERLFDAAMLASGFD 61
D L MRG+++ EINP H I++ L ++ E + E T++ + L+D A+++SGF
Sbjct 662 DTSSLEFMRGRRILEINPDHPIIKDLNAACKNAPEST---EATRVVDLLYDTAIISSGFT 718
Query 62 VEQPAQLAAVLYKGLASQVG 81
+ PA+L +Y+ +A VG
Sbjct 719 PDSPAELGNKIYEMMAMAVG 738
> CE05441
Length=702
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 39/109 (35%), Positives = 59/109 (54%), Gaps = 8/109 (7%)
Query 2 DPMHLRLMRGQKVFEINPSHRIMQQLLQRVR-DGGEDSMKDEDTQLAERLFDAAMLASGF 60
D + M +K EINP H IM+ L RV D + ++KD L LF+ A+LASGF
Sbjct 591 DSSTMGYMAAKKHLEINPDHAIMKTLRDRVEVDKNDKTVKD----LVVLLFETALLASGF 646
Query 61 DVEQPAQLAAVLYKGLASQVGVDPADPINENIDLPEE-EADKTEENAEE 108
+E+P A+ +Y+ + ++G+D D E+ +P A+ E AEE
Sbjct 647 SLEEPQSHASRIYRMI--KLGLDIGDDEIEDSAVPSSCTAEAKIEGAEE 693
> Hs18549738
Length=118
Score = 50.8 bits (120), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 28/79 (35%), Positives = 42/79 (53%), Gaps = 4/79 (5%)
Query 9 MRGQKVFEINPSHRIMQQLLQRVRDGGEDSMKDEDTQLAERLFDAAMLASGFDVEQPAQL 68
M +K EINP H I++ L Q+ S++D L L+ A+L+SGF +E P
Sbjct 19 MAAKKHLEINPDHSIIETLRQKAEADKYKSVQD----LVVLLYKTALLSSGFGLEDPQTH 74
Query 69 AAVLYKGLASQVGVDPADP 87
A +Y+ + VG+D DP
Sbjct 75 ANRIYRMIKLGVGIDENDP 93
> Hs13129150
Length=732
Score = 50.4 bits (119), Expect = 7e-07, Method: Composition-based stats.
Identities = 34/107 (31%), Positives = 54/107 (50%), Gaps = 11/107 (10%)
Query 9 MRGQKVFEINPSHRIMQQLLQRVR-DGGEDSMKDEDTQLAERLFDAAMLASGFDVEQPAQ 67
M +K EINP H I++ L Q+ D + S+KD L L++ A+L+SGF +E P
Sbjct 628 MAAKKHLEINPDHSIIETLRQKAEADKNDKSVKD----LVILLYETALLSSGFSLEDPQT 683
Query 68 LAAVLYKGLASQVGVDPADPINENI------DLPEEEADKTEENAEE 108
A +Y+ + +G+D DP ++ ++P E D EE
Sbjct 684 HANRIYRMIKLGLGIDEDDPTADDTSAAVTEEMPPLEGDDDTSRMEE 730
> At3g07770
Length=803
Score = 50.1 bits (118), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 26/73 (35%), Positives = 43/73 (58%), Gaps = 5/73 (6%)
Query 2 DPMHLRLMRGQKVFEINPSHRIMQQLLQRVRDGGEDSMKDEDTQLA-ERLFDAAMLASGF 60
D + L M+G++VFEINP H I ++ + + DED A + ++DAA+++SGF
Sbjct 689 DTISLDYMKGRRVFEINPDHSI----IKNINAAYNSNPNDEDAMRAIDLMYDAALVSSGF 744
Query 61 DVEQPAQLAAVLY 73
+ PA+L +Y
Sbjct 745 TPDNPAELGGKIY 757
> At5g56000
Length=699
Score = 50.1 bits (118), Expect = 1e-06, Method: Composition-based stats.
Identities = 31/103 (30%), Positives = 55/103 (53%), Gaps = 7/103 (6%)
Query 9 MRGQKVFEINPSHRIMQQLLQRVR-DGGEDSMKDEDTQLAERLFDAAMLASGFDVEQPAQ 67
M +K EINP + IM +L +R D + S+KD L LF+ A+L SGF +++P
Sbjct 599 MSSKKTMEINPENSIMDELRKRAEADKNDKSVKD----LVLLLFETALLTSGFSLDEPNT 654
Query 68 LAAVLYKGLASQVGVDPADPINENIDLP--EEEADKTEENAEE 108
+ +++ L + ++ D + + ++P E++AD EE
Sbjct 655 FGSRIHRMLKLGLSIEEDDAVEADAEMPPLEDDADAEGSKMEE 697
> 7292327
Length=717
Score = 49.7 bits (117), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 28/89 (31%), Positives = 50/89 (56%), Gaps = 5/89 (5%)
Query 2 DPMHLRLMRGQKVFEINPSHRIMQQLLQRV-RDGGEDSMKDEDTQLAERLFDAAMLASGF 60
D + M G+K EINP H I++ L Q+ D + ++KD L LF+ ++L+SGF
Sbjct 606 DTATMGYMAGKKQLEINPDHPIVETLRQKADADKNDKAVKD----LVILLFETSLLSSGF 661
Query 61 DVEQPAQLAAVLYKGLASQVGVDPADPIN 89
++ P A+ +Y+ + +G+D +P+
Sbjct 662 SLDSPQVHASRIYRMIKLGLGIDEDEPMT 690
> YMR186w
Length=705
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 25/74 (33%), Positives = 46/74 (62%), Gaps = 6/74 (8%)
Query 12 QKVFEINPSHRIMQQLLQRVRDGG--EDSMKDEDTQLAERLFDAAMLASGFDVEQPAQLA 69
+K FEI+P I+++L +RV +GG + ++KD L LF+ A+L SGF +E+P A
Sbjct 606 KKTFEISPKSPIIKELKKRVDEGGAQDKTVKD----LTNLLFETALLTSGFSLEEPTSFA 661
Query 70 AVLYKGLASQVGVD 83
+ + + ++ + +D
Sbjct 662 SRINRLISLGLNID 675
> 7301648
Length=787
Score = 47.4 bits (111), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 27/82 (32%), Positives = 46/82 (56%), Gaps = 5/82 (6%)
Query 2 DPMHLRLMRGQKVFEINPSHRIMQQLLQRVR-DGGEDSMKDEDTQLAERLFDAAMLASGF 60
DP + +K EINP H +M++LL+RV D +D+ KD +A +F A L SG+
Sbjct 669 DPQRTYYLNQKKTLEINPRHPLMRELLRRVEADEADDTAKD----MAVMMFRTATLRSGY 724
Query 61 DVEQPAQLAAVLYKGLASQVGV 82
+++ +Q A + + + +GV
Sbjct 725 MLQETSQFADSIEQMMRQTLGV 746
> YPL240c
Length=709
Score = 46.2 bits (108), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 23/74 (31%), Positives = 47/74 (63%), Gaps = 6/74 (8%)
Query 12 QKVFEINPSHRIMQQLLQRVRDGG--EDSMKDEDTQLAERLFDAAMLASGFDVEQPAQLA 69
+K FEI+P I+++L +RV +GG + ++KD L + L++ A+L SGF +++P A
Sbjct 610 KKTFEISPKSPIIKELKKRVDEGGAQDKTVKD----LTKLLYETALLTSGFSLDEPTSFA 665
Query 70 AVLYKGLASQVGVD 83
+ + + ++ + +D
Sbjct 666 SRINRLISLGLNID 679
> Hs16157901
Length=249
Score = 45.8 bits (107), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 27/76 (35%), Positives = 41/76 (53%), Gaps = 5/76 (6%)
Query 9 MRGQKVFEINPSHRIMQQLLQRVR-DGGEDSMKDEDTQLAERLFDAAMLASGFDVEQPAQ 67
M +K EINP H IM+ L Q+ D + ++KD L LF+ A+L+SGF +E P
Sbjct 145 MMAKKHLEINPDHPIMETLRQKAEADKNDKAVKD----LVVLLFETALLSSGFSLEDPQT 200
Query 68 LAAVLYKGLASQVGVD 83
+ +Y + +G D
Sbjct 201 HSNHIYHMIKLGLGTD 216
> Hs20149594
Length=724
Score = 45.4 bits (106), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 26/76 (34%), Positives = 43/76 (56%), Gaps = 5/76 (6%)
Query 9 MRGQKVFEINPSHRIMQQLLQRVR-DGGEDSMKDEDTQLAERLFDAAMLASGFDVEQPAQ 67
M +K EINP H I++ L Q+ D + ++KD L LF+ A+L+SGF +E P
Sbjct 620 MMAKKHLEINPDHPIVETLRQKAEADKNDKAVKD----LVVLLFETALLSSGFSLEDPQT 675
Query 68 LAAVLYKGLASQVGVD 83
+ +Y+ + +G+D
Sbjct 676 HSNRIYRMIKLGLGID 691
> Hs22041746
Length=1595
Score = 44.7 bits (104), Expect = 4e-05, Method: Composition-based stats.
Identities = 31/110 (28%), Positives = 50/110 (45%), Gaps = 11/110 (10%)
Query 9 MRGQKVFEINPSHRIMQQLLQRVR-DGGEDSMKDEDTQLAERLFDAAMLASGFDVEQPAQ 67
M +K EINP H + L Q+ D + S+KD L L++ A+L+S F +E P
Sbjct 926 MAAKKHLEINPDHSFIDTLRQKAETDKNDKSVKD----LVILLYETALLSSDFGLEGPQT 981
Query 68 LAAVLYKGLASQVGVDPADPINENI------DLPEEEADKTEENAEEPMD 111
A +Y+ +G D DP ++ ++P E D E ++
Sbjct 982 HANRIYRMNKLGLGTDEDDPTADDTSAAVTEEMPPLEGDDDTSRMENDVE 1031
> SPAC926.04c
Length=704
Score = 44.7 bits (104), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 34/105 (32%), Positives = 58/105 (55%), Gaps = 8/105 (7%)
Query 9 MRGQKVFEINPSHRIMQQLLQRV-RDGGED-SMKDEDTQLAERLFDAAMLASGFDVEQPA 66
M +K FEINP I+ +L ++V +G ED S+KD LA L++ A+L+SGF ++ P+
Sbjct 603 MSSRKTFEINPKSPIIAELKKKVEENGAEDRSVKD----LATILYETALLSSGFTLDDPS 658
Query 67 QLAAVLYKGLASQVGVDPADPINENIDLPEEEADKTEENAEEPMD 111
A + + ++ +G+ + I+ E+ E NAE M+
Sbjct 659 AYAQRINRLIS--LGLSIDEEEEAPIEEISTESVAAENNAESKME 701
> CE06362
Length=760
Score = 44.3 bits (103), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 28/79 (35%), Positives = 40/79 (50%), Gaps = 7/79 (8%)
Query 2 DPMHLRLMRGQKVFEINPSHRIMQQLLQRVRDGGEDSMKDEDTQLAERLFDAAMLASGFD 61
DP +K FEINP H ++++LL+RV ED+ +L LF+ A L SGF
Sbjct 649 DPTQDFYATQKKTFEINPRHPVIKELLKRVTASEEDTTAASTAKL---LFETATLRSGFS 705
Query 62 VEQPAQLA----AVLYKGL 76
++ A AVL + L
Sbjct 706 LQDQVGFADRIEAVLRQSL 724
> At5g56010
Length=699
Score = 42.7 bits (99), Expect = 2e-04, Method: Composition-based stats.
Identities = 33/103 (32%), Positives = 55/103 (53%), Gaps = 7/103 (6%)
Query 9 MRGQKVFEINPSHRIMQQLLQRV-RDGGEDSMKDEDTQLAERLFDAAMLASGFDVEQPAQ 67
M +K EINP + IM +L +R D + S+KD L LF+ A+L SGF +++P
Sbjct 599 MSSKKTMEINPENSIMDELRKRADADKNDKSVKD----LVLLLFETALLTSGFSLDEPNT 654
Query 68 LAAVLYKGLASQVGVDPADPINENIDLP--EEEADKTEENAEE 108
+ +++ L + +D D + + D+P E++AD EE
Sbjct 655 FGSRIHRMLKLGLSIDDDDVVEADADMPPLEDDADAEGSKMEE 697
> At5g56030
Length=699
Score = 42.7 bits (99), Expect = 2e-04, Method: Composition-based stats.
Identities = 32/103 (31%), Positives = 55/103 (53%), Gaps = 7/103 (6%)
Query 9 MRGQKVFEINPSHRIMQQLLQRV-RDGGEDSMKDEDTQLAERLFDAAMLASGFDVEQPAQ 67
M +K EINP + IM +L +R D + S+KD L LF+ A+L SGF +++P
Sbjct 599 MSSKKTMEINPENSIMDELRKRADADKNDKSVKD----LVLLLFETALLTSGFSLDEPNT 654
Query 68 LAAVLYKGLASQVGVDPADPINENIDLP--EEEADKTEENAEE 108
+ +++ L + +D D + + ++P E++AD EE
Sbjct 655 FGSRIHRMLKLGLSIDDDDAVEADAEMPPLEDDADAEGSKMEE 697
> Hs17448897
Length=101
Score = 35.8 bits (81), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 21/58 (36%), Positives = 33/58 (56%), Gaps = 5/58 (8%)
Query 9 MRGQKVFEINPSHRIMQQLLQRVR-DGGEDSMKDEDTQLAERLFDAAMLASGFDVEQP 65
M +K EINP+H I++ L Q+ D +++D L LF+ +L+SGF +E P
Sbjct 43 MMAKKHLEINPNHPIVEMLQQKAEADRNSKAVRD----LMVLLFETVLLSSGFFLEDP 96
> Hs4507677
Length=803
Score = 35.4 bits (80), Expect = 0.023, Method: Composition-based stats.
Identities = 21/73 (28%), Positives = 38/73 (52%), Gaps = 3/73 (4%)
Query 12 QKVFEINPSHRIMQQLLQRVRDGGEDSMKDEDTQLAERLFDAAMLASGFDVEQPAQLAAV 71
+K FEINP H +++ +L+R+++ +D LA LF+ A L SG+ +
Sbjct 682 KKTFEINPRHPLIRDMLRRIKEDEDDKTV---LDLAVVLFETATLRSGYLLPDTKAYGDR 738
Query 72 LYKGLASQVGVDP 84
+ + L + +DP
Sbjct 739 IERMLRLSLNIDP 751
> ECU02g1100
Length=690
Score = 31.2 bits (69), Expect = 0.43, Method: Composition-based stats.
Identities = 20/79 (25%), Positives = 44/79 (55%), Gaps = 5/79 (6%)
Query 12 QKVFEINPSHRIMQQLLQRVRDGGEDSMKDEDTQLAERLFDAAMLASGFDVEQPAQLAAV 71
+K+FE+NP+H++++ L+ + D E ++ ++ E F+ ++ +GF + P A
Sbjct 615 KKIFEMNPNHQLVKN-LKALFDSNE---IEKMNRILEVFFETVLIHNGFVLSDPKGFCAN 670
Query 72 LYKGLASQVGVDPADPINE 90
++ L S+ V +P+ E
Sbjct 671 VFDFLCSE-EVRCEEPVEE 688
> Hs17933764
Length=1617
Score = 30.8 bits (68), Expect = 0.65, Method: Composition-based stats.
Identities = 23/71 (32%), Positives = 33/71 (46%), Gaps = 7/71 (9%)
Query 9 MRGQKVFEINPSHRIMQQLLQRVRDGGEDSMKDEDTQLAERLFDAAMLASGFDVEQPAQL 68
MR VF I+P R + + D +DED Q ER+ A L + E+P +
Sbjct 1226 MRKDPVFRISPQSRDAKPNPEEPID------EDEDIQ-TERIRTATALTTSILDEKPVII 1278
Query 69 AAVLYKGLASQ 79
A+ L+K A Q
Sbjct 1279 ASCLHKEYAGQ 1289
> SPAPB2B4.03
Length=411
Score = 30.0 bits (66), Expect = 1.2, Method: Composition-based stats.
Identities = 16/53 (30%), Positives = 32/53 (60%), Gaps = 3/53 (5%)
Query 15 FEINPSHRIMQQLLQRVRDGGEDSMKDEDTQLAERLFDAAMLASGFDVEQPAQ 67
FEIN R +L V + G+++ ++DT+ A+R FD + L++ + ++P +
Sbjct 44 FEINLPRR---TVLSDVSNVGKNNADEKDTKKAKRSFDESNLSTNEEADKPVE 93
> YDL028c
Length=764
Score = 28.9 bits (63), Expect = 2.6, Method: Composition-based stats.
Identities = 24/97 (24%), Positives = 46/97 (47%), Gaps = 4/97 (4%)
Query 24 MQQLLQRVRDGGEDSMKDEDTQLAERLFDAAMLASGFDVEQPAQLAAVLYKGLASQVGVD 83
+ + +R+R+ ++ +D + AER ++++ PA+ A L V
Sbjct 126 LDKSFERIRNLRQNMKEDITAKYAERRSKRFLISNRTTKLGPAKRAMTLTNIFDEDVPNS 185
Query 84 PADPIN--ENIDLPEEEADKTEENAEEPMDNRSAEDI 118
P PIN E ++LP E++ +T N +E +N + I
Sbjct 186 PNQPINARETVELPLEDSHQT--NFKERRENTDYDSI 220
> CE16864
Length=374
Score = 28.5 bits (62), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 15/49 (30%), Positives = 27/49 (55%), Gaps = 1/49 (2%)
Query 20 SHRIMQQLLQRVRDGGEDSMKDEDTQLAERLFDAAMLASGFDVEQPAQL 68
+H I++Q +QR D G + ++DE Q+A + M + F+ Q A +
Sbjct 59 AHPIVEQFVQRALDYGVEKLRDEFRQMA-KYTRPDMTQNAFNANQSANI 106
> CE02541
Length=604
Score = 28.5 bits (62), Expect = 3.3, Method: Composition-based stats.
Identities = 13/38 (34%), Positives = 23/38 (60%), Gaps = 5/38 (13%)
Query 2 DPMHLRLMRGQKVFEINPSHRIMQQLL-----QRVRDG 34
D + L++G++ FE+ HRI QL+ Q++R+G
Sbjct 202 DEFRINLLKGKEQFELTEYHRISVQLVASSPQQQLREG 239
> CE22610
Length=1048
Score = 28.1 bits (61), Expect = 3.8, Method: Composition-based stats.
Identities = 16/40 (40%), Positives = 22/40 (55%), Gaps = 1/40 (2%)
Query 16 EINPSHRIMQQL-LQRVRDGGEDSMKDEDTQLAERLFDAA 54
EI+ HR++ + + GG D MK+ DTQ RL D A
Sbjct 849 EIDGCHRVLTPAEIMNIILGGSDRMKELDTQKLRRLTDRA 888
> Hs21361474
Length=1588
Score = 28.1 bits (61), Expect = 4.4, Method: Composition-based stats.
Identities = 16/43 (37%), Positives = 21/43 (48%), Gaps = 0/43 (0%)
Query 75 GLASQVGVDPADPINENIDLPEEEADKTEENAEEPMDNRSAED 117
G A Q D +P I+ PEE A++ E AEEP + D
Sbjct 1441 GEAEQPNGDADEPDGAGIEDPEERAEEPEGKAEEPEGDADEPD 1483
> Hs10442822_2
Length=4498
Score = 27.7 bits (60), Expect = 4.7, Method: Composition-based stats.
Identities = 12/30 (40%), Positives = 18/30 (60%), Gaps = 0/30 (0%)
Query 83 DPADPINENIDLPEEEADKTEENAEEPMDN 112
DP D I I LP + D E++ EEP+++
Sbjct 492 DPQDTITSLILLPPDILDNREDDCEEPIED 521
> CE23002
Length=313
Score = 27.3 bits (59), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 12/40 (30%), Positives = 21/40 (52%), Gaps = 0/40 (0%)
Query 10 RGQKVFEINPSHRIMQQLLQRVRDGGEDSMKDEDTQLAER 49
R +K+F ++P + + L +R R G ++ DE T R
Sbjct 160 RKKKIFALHPLNGLPSALAERCRSGSAPAINDETTHTWLR 199
> At1g75010
Length=712
Score = 27.3 bits (59), Expect = 7.0, Method: Composition-based stats.
Identities = 27/87 (31%), Positives = 44/87 (50%), Gaps = 11/87 (12%)
Query 39 MKDEDTQLAERLFDAAMLASGFDVEQPAQL----AAVLYKGLAS-QVGVDPADPINENID 93
+KDE+ E + + +M G+ +EQ Q AVL G+ + VGV P+ +N N+
Sbjct 426 LKDENGAEREPIANWSM-DQGYQIEQKWQADSGDTAVLSLGIVNLPVGVRPSKKLNSNLS 484
Query 94 LPEE---EADKTEENAEEPMDNRSAED 117
+ + +AD EE+ P N S +D
Sbjct 485 VASQLSRKADSREESFFNP--NGSTKD 509
> At3g58980
Length=977
Score = 27.3 bits (59), Expect = 7.6, Method: Composition-based stats.
Identities = 15/63 (23%), Positives = 31/63 (49%), Gaps = 0/63 (0%)
Query 8 LMRGQKVFEINPSHRIMQQLLQRVRDGGEDSMKDEDTQLAERLFDAAMLASGFDVEQPAQ 67
L+ + +FE + + ++ ++RVR ED ++D++ + R + L +G Q
Sbjct 760 LVNMKALFEARINFIVTEKQIKRVRAPNEDWLQDDEVDVVLRFSNVVKLMNGIQNVQKLY 819
Query 68 LAA 70
L A
Sbjct 820 LTA 822
Lambda K H
0.313 0.133 0.365
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1180352192
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40