bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1616_orf1
Length=118
Score E
Sequences producing significant alignments: (Bits) Value
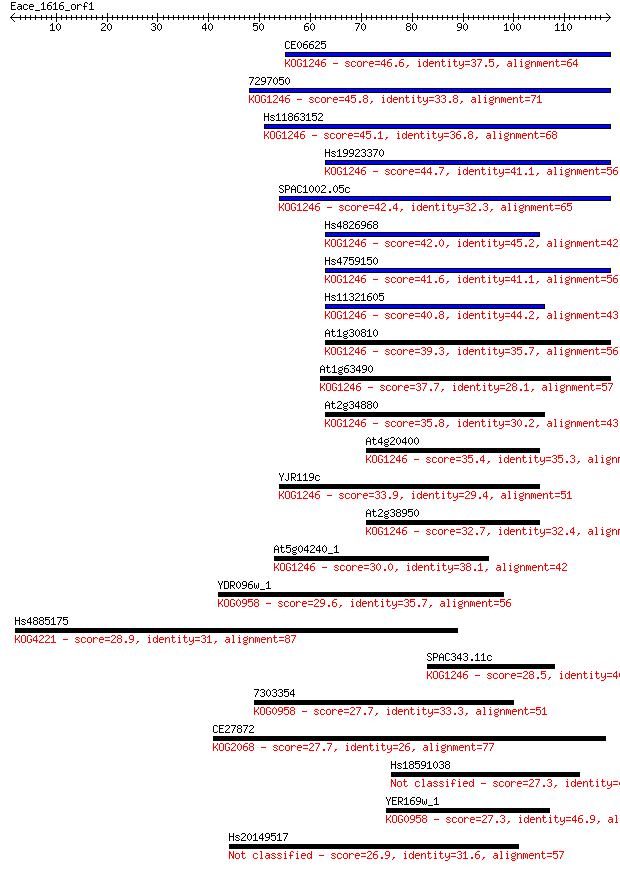
CE06625 46.6 1e-05
7297050 45.8 2e-05
Hs11863152 45.1 3e-05
Hs19923370 44.7 5e-05
SPAC1002.05c 42.4 2e-04
Hs4826968 42.0 3e-04
Hs4759150 41.6 3e-04
Hs11321605 40.8 7e-04
At1g30810 39.3 0.002
At1g63490 37.7 0.005
At2g34880 35.8 0.017
At4g20400 35.4 0.024
YJR119c 33.9 0.070
At2g38950 32.7 0.14
At5g04240_1 30.0 1.2
YDR096w_1 29.6 1.5
Hs4885175 28.9 2.2
SPAC343.11c 28.5 2.8
7303354 27.7 5.5
CE27872 27.7 5.6
Hs18591038 27.3 6.3
YER169w_1 27.3 7.3
Hs20149517 26.9 9.3
> CE06625
Length=1430
Score = 46.6 bits (109), Expect = 1e-05, Method: Composition-based stats.
Identities = 24/66 (36%), Positives = 37/66 (56%), Gaps = 8/66 (12%)
Query 55 PAVPELRFPSRAAFATNPITFFESLRAIGREFGAVKIIPPPDWDPPFAL--EALTNDQAL 112
P + + +P+ F ++PI + +R +FG VKI+PP ++ PPFA+ EA T
Sbjct 53 PPMAPIYYPTEEEF-SDPIEYVAKIRHEAEKFGVVKIVPPANFKPPFAIDKEAFT----- 106
Query 113 FHVRTQ 118
F RTQ
Sbjct 107 FRPRTQ 112
> 7297050
Length=1838
Score = 45.8 bits (107), Expect = 2e-05, Method: Composition-based stats.
Identities = 24/71 (33%), Positives = 37/71 (52%), Gaps = 5/71 (7%)
Query 48 EMNYSSLPAVPELRFPSRAAFATNPITFFESLRAIGREFGAVKIIPPPDWDPPFALEALT 107
E ++ + P P R P+ F NP+ + +R+I + G KI+PP W PPFA++
Sbjct 152 EFHFDTPPECPVFR-PTTEEF-KNPLAYISKIRSIAEKCGIAKILPPATWSPPFAVDV-- 207
Query 108 NDQALFHVRTQ 118
D+ F R Q
Sbjct 208 -DKLRFVPRVQ 217
> Hs11863152
Length=1246
Score = 45.1 bits (105), Expect = 3e-05, Method: Composition-based stats.
Identities = 25/68 (36%), Positives = 39/68 (57%), Gaps = 6/68 (8%)
Query 51 YSSLPAVPELRFPSRAAFATNPITFFESLRAIGREFGAVKIIPPPDWDPPFALEALTNDQ 110
++++ +P LR PS F +P+ + ES+RA +FG ++IPPPDW P E ND+
Sbjct 551 WAAMDEIPVLR-PSAKEFH-DPLIYIESVRAQVEKFGMCRVIPPPDWRP----ECKLNDE 604
Query 111 ALFHVRTQ 118
F + Q
Sbjct 605 MRFVTQIQ 612
> Hs19923370
Length=1681
Score = 44.7 bits (104), Expect = 5e-05, Method: Composition-based stats.
Identities = 23/56 (41%), Positives = 29/56 (51%), Gaps = 4/56 (7%)
Query 63 PSRAAFATNPITFFESLRAIGREFGAVKIIPPPDWDPPFALEALTNDQALFHVRTQ 118
PS FA +P F +R I + G K+ PPPDW PPFA + D+ F R Q
Sbjct 174 PSWEEFA-DPFAFIHKIRPIAEQTGICKVRPPPDWQPPFACDG---DKLHFTPRIQ 225
> SPAC1002.05c
Length=715
Score = 42.4 bits (98), Expect = 2e-04, Method: Composition-based stats.
Identities = 21/65 (32%), Positives = 33/65 (50%), Gaps = 7/65 (10%)
Query 54 LPAVPELRFPSRAAFATNPITFFESLRAIGREFGAVKIIPPPDWDPPFALEALTNDQALF 113
LP P + +P + F + I + + IG ++G +KI+PP W+PP L D F
Sbjct 57 LPVAP-VFYPDKEEF-QDSIGYINKIAPIGEKYGIIKIVPPAGWNPPMQL-----DMNKF 109
Query 114 HVRTQ 118
RT+
Sbjct 110 SFRTR 114
> Hs4826968
Length=1722
Score = 42.0 bits (97), Expect = 3e-04, Method: Composition-based stats.
Identities = 19/42 (45%), Positives = 25/42 (59%), Gaps = 1/42 (2%)
Query 63 PSRAAFATNPITFFESLRAIGREFGAVKIIPPPDWDPPFALE 104
PS F T+P++F +R + + G KI PP DW PPFA E
Sbjct 24 PSWEEF-TDPLSFIGRIRPLAEKTGICKIRPPKDWQPPFACE 64
> Hs4759150
Length=1539
Score = 41.6 bits (96), Expect = 3e-04, Method: Composition-based stats.
Identities = 23/56 (41%), Positives = 29/56 (51%), Gaps = 4/56 (7%)
Query 63 PSRAAFATNPITFFESLRAIGREFGAVKIIPPPDWDPPFALEALTNDQALFHVRTQ 118
PS A F +P+ + +R I + G KI PP DW PPFA+E D F R Q
Sbjct 19 PSWAEF-QDPLGYIAKIRPIAEKSGICKIRPPADWQPPFAVEV---DNFRFTPRVQ 70
> Hs11321605
Length=1560
Score = 40.8 bits (94), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 19/43 (44%), Positives = 25/43 (58%), Gaps = 1/43 (2%)
Query 63 PSRAAFATNPITFFESLRAIGREFGAVKIIPPPDWDPPFALEA 105
PS A F +P+ + +R I + G KI PP DW PPFA+E
Sbjct 19 PSWAEF-RDPLGYIAKIRPIAEKSGICKIRPPADWQPPFAVEV 60
> At1g30810
Length=787
Score = 39.3 bits (90), Expect = 0.002, Method: Composition-based stats.
Identities = 20/57 (35%), Positives = 28/57 (49%), Gaps = 2/57 (3%)
Query 63 PSRAAFATNPITFFESLRAIGREFGAVKIIPPPDWDPPFAL-EALTNDQALFHVRTQ 118
PS F +P+ + E +R + +G +IIPP W PP L E +Q F R Q
Sbjct 64 PSLEEF-VDPLAYIEKIRPLAEPYGICRIIPPSTWKPPCRLKEKSIWEQTKFPTRIQ 119
> At1g63490
Length=1324
Score = 37.7 bits (86), Expect = 0.005, Method: Composition-based stats.
Identities = 16/57 (28%), Positives = 29/57 (50%), Gaps = 4/57 (7%)
Query 62 FPSRAAFATNPITFFESLRAIGREFGAVKIIPPPDWDPPFALEALTNDQALFHVRTQ 118
+P+ F +P+ + ++ +G KI+PP +W PPF L+ + F +TQ
Sbjct 31 YPTEDEF-KDPLEYIHKIKPEAEVYGICKIVPPNNWKPPFGLDL---ESVKFPTKTQ 83
> At2g34880
Length=806
Score = 35.8 bits (81), Expect = 0.017, Method: Composition-based stats.
Identities = 13/43 (30%), Positives = 23/43 (53%), Gaps = 1/43 (2%)
Query 63 PSRAAFATNPITFFESLRAIGREFGAVKIIPPPDWDPPFALEA 105
P+ F + + + E +R + FG +I+PP +W PP L+
Sbjct 66 PTSEEFE-DTLAYIEKIRPLAESFGICRIVPPSNWSPPCRLKG 107
> At4g20400
Length=872
Score = 35.4 bits (80), Expect = 0.024, Method: Composition-based stats.
Identities = 12/34 (35%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 71 NPITFFESLRAIGREFGAVKIIPPPDWDPPFALE 104
+P+ + E LR+ +G +I+PP W PP L+
Sbjct 48 DPLGYIEKLRSKAESYGICRIVPPVAWRPPCPLK 81
> YJR119c
Length=728
Score = 33.9 bits (76), Expect = 0.070, Method: Composition-based stats.
Identities = 15/53 (28%), Positives = 31/53 (58%), Gaps = 4/53 (7%)
Query 54 LPAVPELRFPSRAAFATNPITFFES--LRAIGREFGAVKIIPPPDWDPPFALE 104
+ +P L +P+ F NPI + + ++ +G +G VK++PP + PP +++
Sbjct 1 MEEIPAL-YPTEQEF-KNPIDYLSNPHIKRLGVRYGMVKVVPPNGFCPPLSID 51
> At2g38950
Length=694
Score = 32.7 bits (73), Expect = 0.14, Method: Composition-based stats.
Identities = 11/34 (32%), Positives = 19/34 (55%), Gaps = 0/34 (0%)
Query 71 NPITFFESLRAIGREFGAVKIIPPPDWDPPFALE 104
+ +++ SLR +G ++PPP W PP L+
Sbjct 106 DTLSYISSLRDRAEPYGICCVVPPPSWKPPCLLK 139
> At5g04240_1
Length=1165
Score = 30.0 bits (66), Expect = 1.2, Method: Composition-based stats.
Identities = 16/42 (38%), Positives = 21/42 (50%), Gaps = 2/42 (4%)
Query 53 SLPAVPELRFPSRAAFATNPITFFESLRAIGREFGAVKIIPP 94
+LP P R P+ FA +PI + + FG KIIPP
Sbjct 12 ALPLAPVFR-PTDTEFA-DPIAYISKIEKEASAFGICKIIPP 51
> YDR096w_1
Length=734
Score = 29.6 bits (65), Expect = 1.5, Method: Composition-based stats.
Identities = 20/56 (35%), Positives = 29/56 (51%), Gaps = 6/56 (10%)
Query 42 LETRRVEMNYSSLPAVPELRFPSRAAFATNPITFFESLRAIGREFGAVKIIPPPDW 97
+E + VE+ + VP + PS FA N F + + G E G VK+IPP +W
Sbjct 1 MEIKPVEV----IDGVPVFK-PSMMEFA-NFQYFIDEITKFGIENGIVKVIPPKEW 50
> Hs4885175
Length=1447
Score = 28.9 bits (63), Expect = 2.2, Method: Composition-based stats.
Identities = 27/90 (30%), Positives = 42/90 (46%), Gaps = 10/90 (11%)
Query 2 AGATQAAGERPQRARRAPR---MFEVDAAADDKLLKVAIGRSLLETRRVEMNYSSLPAVP 58
AG++ + ER ARRAPR M +DA +++ + AI LE+ + Y + P
Sbjct 1213 AGSSMSTLERSLAARRAPRAKLMIPMDAQSNNPAVVSAIPVPTLESAQ----YPGILPSP 1268
Query 59 ELRFPSRAAFATNPITFFESLRAIGREFGA 88
+P F P+ F ++ R FGA
Sbjct 1269 TCGYP-HPQFTLRPVPF--PTLSVDRGFGA 1295
> SPAC343.11c
Length=1588
Score = 28.5 bits (62), Expect = 2.8, Method: Composition-based stats.
Identities = 10/25 (40%), Positives = 15/25 (60%), Gaps = 0/25 (0%)
Query 83 GREFGAVKIIPPPDWDPPFALEALT 107
R+ GAV++IPP +W P + T
Sbjct 107 ARKSGAVRVIPPDNWKCPLTINTTT 131
> 7303354
Length=717
Score = 27.7 bits (60), Expect = 5.5, Method: Composition-based stats.
Identities = 17/53 (32%), Positives = 26/53 (49%), Gaps = 5/53 (9%)
Query 49 MNYSSLPAVPELRFPSRAAFATNP--ITFFESLRAIGREFGAVKIIPPPDWDP 99
M S +P + R P+ F P + + ES A + G K++PPP+W P
Sbjct 1 MKMSEVPRIKVFR-PTWEEFKDFPKYVAYMESQGA--HKAGLAKVVPPPEWVP 50
> CE27872
Length=796
Score = 27.7 bits (60), Expect = 5.6, Method: Composition-based stats.
Identities = 20/77 (25%), Positives = 33/77 (42%), Gaps = 6/77 (7%)
Query 41 LLETRRVEMNYSSLPAVPELRFPSRAAFATNPITFFESLRAIGREFGAVKIIPPPDWDPP 100
L T E +Y +PA P++ A P+ +E+L + I+ P P
Sbjct 389 LARTSFSENDYLGIPA------PAKHQEAPAPLMQWEALLGLSSPSAQSTIVEPSSLFPT 442
Query 101 FALEALTNDQALFHVRT 117
F +++ N Q+LF T
Sbjct 443 FKMDSGFNSQSLFGTHT 459
> Hs18591038
Length=201
Score = 27.3 bits (59), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 17/37 (45%), Positives = 20/37 (54%), Gaps = 5/37 (13%)
Query 76 FESLRAIGREFGAVKIIPPPDWDPPFALEALTNDQAL 112
FESL AI +G ++ P P ALEA T D AL
Sbjct 74 FESLAAIRFYYGPARVCP-----RPLALEARTTDWAL 105
> YER169w_1
Length=706
Score = 27.3 bits (59), Expect = 7.3, Method: Composition-based stats.
Identities = 15/39 (38%), Positives = 22/39 (56%), Gaps = 7/39 (17%)
Query 75 FFESLRAI---GREFGAVKIIPPPDW----DPPFALEAL 106
F+ +AI G + G VK+IPP +W D P++ E L
Sbjct 27 FYAYCKAINKYGMKSGVVKVIPPKEWKDKLDLPYSAETL 65
> Hs20149517
Length=377
Score = 26.9 bits (58), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 18/57 (31%), Positives = 25/57 (43%), Gaps = 5/57 (8%)
Query 44 TRRVEMNYSSLPAVPELRFPSRAAFATNPITFFESLRAIGREFGAVKIIPPPDWDPP 100
+R E + LPAVP + +A F P T E+ R G G+ P +W P
Sbjct 158 SREGEAHELRLPAVPSINGTFQADFPLGPATHGETYRCFGSFHGS-----PYEWSDP 209
Lambda K H
0.321 0.135 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1167969826
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40