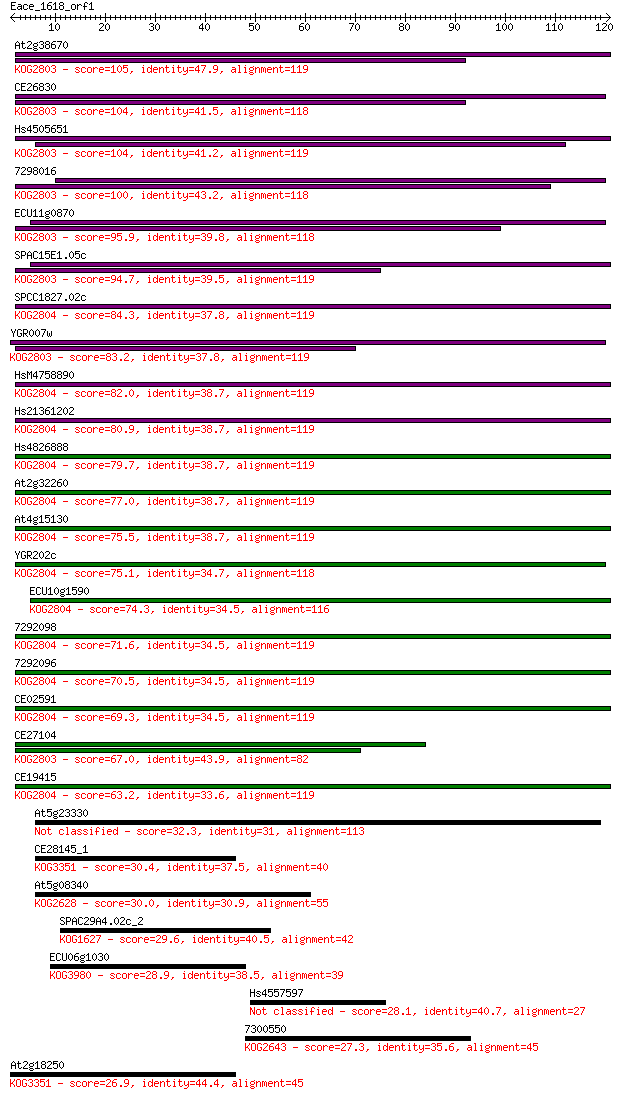
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1618_orf1
Length=120
Score E
Sequences producing significant alignments: (Bits) Value
At2g38670 105 2e-23
CE26830 104 5e-23
Hs4505651 104 5e-23
7298016 100 9e-22
ECU11g0870 95.9 2e-20
SPAC15E1.05c 94.7 4e-20
SPCC1827.02c 84.3 5e-17
YGR007w 83.2 1e-16
HsM4758890 82.0 2e-16
Hs21361202 80.9 6e-16
Hs4826888 79.7 1e-15
At2g32260 77.0 9e-15
At4g15130 75.5 2e-14
YGR202c 75.1 3e-14
ECU10g1590 74.3 5e-14
7292098 71.6 4e-13
7292096 70.5 7e-13
CE02591 69.3 1e-12
CE27104 67.0 8e-12
CE19415 63.2 1e-10
At5g23330 32.3 0.19
CE28145_1 30.4 0.84
At5g08340 30.0 1.0
SPAC29A4.02c_2 29.6 1.3
ECU06g1030 28.9 2.5
Hs4557597 28.1 4.6
7300550 27.3 6.2
At2g18250 26.9 8.7
> At2g38670
Length=421
Score = 105 bits (262), Expect = 2e-23, Method: Composition-based stats.
Identities = 57/123 (46%), Positives = 71/123 (57%), Gaps = 5/123 (4%)
Query 2 VQTDGVFDLLHSGHLNALRQARELGCCLVVGINGDEAVLAAKGFLPVYSAAERAEVIASC 61
V DG FD++H GH NALRQAR LG LVVG+ DE ++A KG PV ER ++ +
Sbjct 57 VYMDGCFDMMHYGHCNALRQARALGDQLVVGVVSDEEIIANKG-PPVTPLHERMTMVKAV 115
Query 62 KWVDEVVVGTPYAVS----THLLDALSCEFAAHGDDWVCSSTGEDAYAGPRAAGRMKTFA 117
KWVDEV+ PYA++ L D ++ HGDD G DAYA + AGR K
Sbjct 116 KWVDEVISDAPYAITEDFMKKLFDEYQIDYIIHGDDPCVLPDGTDAYALAKKAGRYKQIK 175
Query 118 RTE 120
RTE
Sbjct 176 RTE 178
Score = 80.5 bits (197), Expect = 8e-16, Method: Composition-based stats.
Identities = 37/91 (40%), Positives = 56/91 (61%), Gaps = 1/91 (1%)
Query 2 VQTDGVFDLLHSGHLNALRQARELGCCLVVGINGDEAVLAAKG-FLPVYSAAERAEVIAS 60
+ DG FDL H+GH+ LR+ARELG L+VGI+ D+ V A +G P+ + ER+ + +
Sbjct 257 IYIDGAFDLFHAGHVEILRRARELGDFLLVGIHNDQTVSAKRGAHRPIMNLHERSLSVLA 316
Query 61 CKWVDEVVVGTPYAVSTHLLDALSCEFAAHG 91
C++VDEV++G P+ VS + HG
Sbjct 317 CRYVDEVIIGAPWEVSRDTITTFDISLVVHG 347
> CE26830
Length=370
Score = 104 bits (259), Expect = 5e-23, Method: Composition-based stats.
Identities = 49/118 (41%), Positives = 69/118 (58%), Gaps = 1/118 (0%)
Query 2 VQTDGVFDLLHSGHLNALRQARELGCCLVVGINGDEAVLAAKGFLPVYSAAERAEVIASC 61
V DG +D++H GH N LRQA++ G L+VG++ DE + KG PV++ ER ++A
Sbjct 15 VWADGCYDMVHFGHANQLRQAKQFGQKLIVGVHNDEEIRLHKG-PPVFNEQERYRMVAGI 73
Query 62 KWVDEVVVGTPYAVSTHLLDALSCEFAAHGDDWVCSSTGEDAYAGPRAAGRMKTFART 119
KWVDEVV PYA + LD C+F HGDD ++ G+D Y + R + RT
Sbjct 74 KWVDEVVENAPYATTVETLDKYDCDFCVHGDDITLTADGKDTYQEVKDHQRYRECKRT 131
Score = 63.9 bits (154), Expect = 6e-11, Method: Composition-based stats.
Identities = 36/91 (39%), Positives = 51/91 (56%), Gaps = 1/91 (1%)
Query 2 VQTDGVFDLLHSGHLNALRQARELGCCLVVGINGDEAVLAAKGF-LPVYSAAERAEVIAS 60
V G FDL H GHL L +A+E G L+VGI D+ V KG P+ S ER + +
Sbjct 206 VYVTGSFDLFHIGHLAFLEKAKEFGDYLIVGILSDQTVNQYKGSNHPIMSIHERVLSVLA 265
Query 61 CKWVDEVVVGTPYAVSTHLLDALSCEFAAHG 91
K V+EVV G PY +++ +LD + + +G
Sbjct 266 YKPVNEVVFGAPYEITSDILDQFNVQAVING 296
> Hs4505651
Length=389
Score = 104 bits (259), Expect = 5e-23, Method: Composition-based stats.
Identities = 49/119 (41%), Positives = 71/119 (59%), Gaps = 1/119 (0%)
Query 2 VQTDGVFDLLHSGHLNALRQARELGCCLVVGINGDEAVLAAKGFLPVYSAAERAEVIASC 61
V DG +D++H GH N LRQAR +G L+VG++ DE + KG PV++ ER +++ +
Sbjct 25 VWCDGCYDMVHYGHSNQLRQARAMGDYLIVGVHTDEEIAKHKG-PPVFTQEERYKMVQAI 83
Query 62 KWVDEVVVGTPYAVSTHLLDALSCEFAAHGDDWVCSSTGEDAYAGPRAAGRMKTFARTE 120
KWVDEVV PY + LD +C+F HG+D + G D Y + AGR + RT+
Sbjct 84 KWVDEVVPAAPYVTTLETLDKYNCDFCVHGNDITLTVDGRDTYEEVKQAGRYRECKRTQ 142
Score = 63.5 bits (153), Expect = 8e-11, Method: Composition-based stats.
Identities = 36/110 (32%), Positives = 56/110 (50%), Gaps = 4/110 (3%)
Query 6 GVFDLLHSGHLNALRQARELG--CCLVVGINGDEAVLAAKG-FLPVYSAAERAEVIASCK 62
G FDL H GH++ L + L ++ G++ D+ V KG P+ + ER + +C+
Sbjct 220 GAFDLFHIGHVDFLEKVHRLAERPYIIAGLHFDQEVNHYKGKNYPIMNLHERTLSVLACR 279
Query 63 WVDEVVVGTPYAVSTHLLDALSCEFAAHGD-DWVCSSTGEDAYAGPRAAG 111
+V EVV+G PYAV+ LL + HG + + G D Y P+ G
Sbjct 280 YVSEVVIGAPYAVTAELLSHFKVDLVCHGKTEIIPDRDGSDPYQEPKRRG 329
> 7298016
Length=363
Score = 100 bits (248), Expect = 9e-22, Method: Composition-based stats.
Identities = 47/110 (42%), Positives = 69/110 (62%), Gaps = 1/110 (0%)
Query 10 LLHSGHLNALRQARELGCCLVVGINGDEAVLAAKGFLPVYSAAERAEVIASCKWVDEVVV 69
++H GH N+LRQA+ LG ++VGI+ DE + KG PV++ ER +++ KWVDEVV+
Sbjct 1 MVHFGHANSLRQAKALGDKVIVGIHTDEEITKHKG-PPVFTEEERVKMVKGIKWVDEVVL 59
Query 70 GTPYAVSTHLLDALSCEFAAHGDDWVCSSTGEDAYAGPRAAGRMKTFART 119
G PY + +LD +C+F HGDD ++ G D Y ++A R K RT
Sbjct 60 GAPYVTTLDVLDQNNCDFCVHGDDITMTAEGVDTYHLVKSANRYKEVKRT 109
Score = 79.0 bits (193), Expect = 2e-15, Method: Composition-based stats.
Identities = 42/109 (38%), Positives = 63/109 (57%), Gaps = 2/109 (1%)
Query 2 VQTDGVFDLLHSGHLNALRQARELGCCLVVGINGDEAVLAAKGF-LPVYSAAERAEVIAS 60
V G FDL H GHL+ L +A++LG L+VG++ D V + KG P+ + ER + +
Sbjct 197 VYVAGAFDLFHVGHLDFLEKAKKLGDYLIVGLHTDPVVNSYKGSNYPIMNLHERVLSVLA 256
Query 61 CKWVDEVVVGTPYAVSTHLLDALSCEFAAHGDDWVCSSTGE-DAYAGPR 108
CK+V+EVV+G PY V+ LL+ + HG + G+ D YA P+
Sbjct 257 CKFVNEVVIGAPYCVTEELLEHFKIDVVCHGRTPIALENGKIDPYAVPK 305
> ECU11g0870
Length=322
Score = 95.9 bits (237), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 46/115 (40%), Positives = 65/115 (56%), Gaps = 1/115 (0%)
Query 5 DGVFDLLHSGHLNALRQARELGCCLVVGINGDEAVLAAKGFLPVYSAAERAEVIASCKWV 64
DG FD+ H GH NALRQ++ LG L+ G++ ++ KG LPV ER EV+ C++V
Sbjct 12 DGCFDMFHYGHANALRQSKALGDYLIAGVHSSLSINQEKG-LPVMEDEERYEVVEGCRYV 70
Query 65 DEVVVGTPYAVSTHLLDALSCEFAAHGDDWVCSSTGEDAYAGPRAAGRMKTFART 119
DEVV P+ T ++ AHG+D V S+G+D+Y R G + RT
Sbjct 71 DEVVRDAPFVTQTSMIKEYGVSIVAHGNDIVLDSSGQDSYCQVRRMGIFREVERT 125
Score = 68.6 bits (166), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 39/97 (40%), Positives = 53/97 (54%), Gaps = 8/97 (8%)
Query 2 VQTDGVFDLLHSGHLNALRQARELGCCLVVGINGDEAVLAAKGFLPVYSAAERAEVIASC 61
V DG FDL H+GH+ +LR AR +G L+VGI+ DE PV S ER + +C
Sbjct 180 VFMDGNFDLFHAGHVASLRIARGMGDYLIVGIHDDETTKEYTRSYPVLSTKERMLTLMAC 239
Query 62 KWVDEVVVGTPYAVSTHLLDALSCEFAAHGDDWVCSS 98
++VDE+VV +PY V + + HG D V S
Sbjct 240 RYVDEIVV-SPYLVGSEFI-------KRHGIDVVAPS 268
> SPAC15E1.05c
Length=365
Score = 94.7 bits (234), Expect = 4e-20, Method: Composition-based stats.
Identities = 47/116 (40%), Positives = 62/116 (53%), Gaps = 1/116 (0%)
Query 5 DGVFDLLHSGHLNALRQARELGCCLVVGINGDEAVLAAKGFLPVYSAAERAEVIASCKWV 64
DG D H GH NA+ QA++LG LV+GI+ DE + KG PV + ER +CKWV
Sbjct 14 DGCMDFFHYGHSNAILQAKQLGETLVIGIHSDEEITLNKG-PPVMTLEERCLSANTCKWV 72
Query 65 DEVVVGTPYAVSTHLLDALSCEFAAHGDDWVCSSTGEDAYAGPRAAGRMKTFARTE 120
DEVV PY + C++ HGDD + G+D Y +AA + RTE
Sbjct 73 DEVVPSAPYVFDLEWMRRYGCQYVVHGDDISTDANGDDCYRFAKAADQYLEVKRTE 128
Score = 33.9 bits (76), Expect = 0.076, Method: Composition-based stats.
Identities = 21/75 (28%), Positives = 35/75 (46%), Gaps = 6/75 (8%)
Query 2 VQTDGVFDLLHSGHLNALRQAREL--GCCLVVGINGDEAVLAAKGFLPVYSAAERAEVIA 59
+ DG +DL H++AL + G ++ GI DE P+ + ER +
Sbjct 205 IYIDGDWDLFTEKHISALELCTRMFPGIPIMAGIFADEKCFEK----PMLNLLERILNLL 260
Query 60 SCKWVDEVVVGTPYA 74
CK++ ++VG P A
Sbjct 261 QCKYISSILVGPPPA 275
> SPCC1827.02c
Length=354
Score = 84.3 bits (207), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 45/121 (37%), Positives = 62/121 (51%), Gaps = 3/121 (2%)
Query 2 VQTDGVFDLLHSGHLNALRQAREL--GCCLVVGINGDEAVLAAKGFLPVYSAAERAEVIA 59
V DGVFDL H GH+ L QA+++ L+VG+ D+ KG L V + ERAE +
Sbjct 96 VYADGVFDLFHIGHMRQLEQAKKVFPNVHLIVGLPNDQLTHRLKG-LTVMNDKERAEALR 154
Query 60 SCKWVDEVVVGTPYAVSTHLLDALSCEFAAHGDDWVCSSTGEDAYAGPRAAGRMKTFART 119
CKWVDEV+ P+ ++ L+ +F AH D S D Y + G+ RT
Sbjct 155 HCKWVDEVLENAPWVITPEFLEEHKIDFVAHDDIPYASDDSGDIYLPVKKVGKFIPTKRT 214
Query 120 E 120
E
Sbjct 215 E 215
> YGR007w
Length=323
Score = 83.2 bits (204), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 45/123 (36%), Positives = 59/123 (47%), Gaps = 5/123 (4%)
Query 1 AVQTDGVFDLLHSGHLNALRQAREL----GCCLVVGINGDEAVLAAKGFLPVYSAAERAE 56
V DG FD H GH A+ QAR L G++ DE + KG PV +++ER E
Sbjct 9 KVWIDGCFDFTHHGHAGAILQARRTVSKENGKLFCGVHTDEDIQHNKG-TPVMNSSERYE 67
Query 57 VIASCKWVDEVVVGTPYAVSTHLLDALSCEFAAHGDDWVCSSTGEDAYAGPRAAGRMKTF 116
S +W EVV PY + +D C++ HGDD + GED Y + GR K
Sbjct 68 HTRSNRWCSEVVEAAPYVTDPNWMDKYQCQYVVHGDDITIDANGEDCYKLVKEMGRFKVV 127
Query 117 ART 119
RT
Sbjct 128 KRT 130
Score = 35.8 bits (81), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 22/71 (30%), Positives = 34/71 (47%), Gaps = 9/71 (12%)
Query 2 VQTDGVFDLLHSGHLNALRQAR---ELGCCLVVGINGDEAVLAAKGFLPVYSAAERAEVI 58
V DG FDL H G ++ LR+ + L+VGI + + + ER +
Sbjct 200 VYVDGDFDLFHMGDIDQLRKLKMDLHPDKKLIVGITTSDYSST------IMTMKERVLSV 253
Query 59 ASCKWVDEVVV 69
SCK+VD V++
Sbjct 254 LSCKYVDAVII 264
> HsM4758890
Length=330
Score = 82.0 bits (201), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 46/121 (38%), Positives = 64/121 (52%), Gaps = 3/121 (2%)
Query 2 VQTDGVFDLLHSGHLNALRQAREL--GCCLVVGINGDEAVLAAKGFLPVYSAAERAEVIA 59
V DG+FDL HSGH AL QA+ L L+VG+ D+ KGF V + AER E +
Sbjct 79 VYADGIFDLFHSGHARALMQAKTLFPNSYLLVGVCSDDLTHKFKGFT-VMNEAERYEALR 137
Query 60 SCKWVDEVVVGTPYAVSTHLLDALSCEFAAHGDDWVCSSTGEDAYAGPRAAGRMKTFART 119
C++VDEV+ P+ ++ L+ +F AH D S+ +D Y + AG RT
Sbjct 138 HCRYVDEVIRDAPWTLTPEFLEKHKIDFVAHDDIPYSSAGSDDVYKHIKEAGMFVPTQRT 197
Query 120 E 120
E
Sbjct 198 E 198
> Hs21361202
Length=369
Score = 80.9 bits (198), Expect = 6e-16, Method: Composition-based stats.
Identities = 46/121 (38%), Positives = 64/121 (52%), Gaps = 3/121 (2%)
Query 2 VQTDGVFDLLHSGHLNALRQAREL--GCCLVVGINGDEAVLAAKGFLPVYSAAERAEVIA 59
V DG+FDL HSGH AL QA+ L L+VG+ D+ KGF V + AER E +
Sbjct 79 VYADGIFDLFHSGHARALMQAKTLFPNSYLLVGVCSDDLTHKFKGF-TVMNEAERYEALR 137
Query 60 SCKWVDEVVVGTPYAVSTHLLDALSCEFAAHGDDWVCSSTGEDAYAGPRAAGRMKTFART 119
C++VDEV+ P+ ++ L+ +F AH D S+ +D Y + AG RT
Sbjct 138 HCRYVDEVIRDAPWTLTPEFLEKHKIDFVAHDDIPYSSAGSDDVYKHIKEAGMFVPTQRT 197
Query 120 E 120
E
Sbjct 198 E 198
> Hs4826888
Length=367
Score = 79.7 bits (195), Expect = 1e-15, Method: Composition-based stats.
Identities = 46/121 (38%), Positives = 62/121 (51%), Gaps = 3/121 (2%)
Query 2 VQTDGVFDLLHSGHLNALRQAREL--GCCLVVGINGDEAVLAAKGFLPVYSAAERAEVIA 59
V DG+FDL HSGH AL QA+ L L+VG+ DE KGF V + ER + +
Sbjct 79 VYADGIFDLFHSGHARALMQAKNLFPNTYLIVGVCSDELTHNFKGF-TVMNENERYDAVQ 137
Query 60 SCKWVDEVVVGTPYAVSTHLLDALSCEFAAHGDDWVCSSTGEDAYAGPRAAGRMKTFART 119
C++VDEVV P+ ++ L +F AH D S+ +D Y + AG RT
Sbjct 138 HCRYVDEVVRNAPWTLTPEFLAEHRIDFVAHDDIPYSSAGSDDVYKHIKEAGMFAPTQRT 197
Query 120 E 120
E
Sbjct 198 E 198
> At2g32260
Length=332
Score = 77.0 bits (188), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 46/124 (37%), Positives = 61/124 (49%), Gaps = 6/124 (4%)
Query 2 VQTDGVFDLLHSGHLNALRQAR---ELGCCLVVGINGDEAVLAAKGFLPVYSAAERAEVI 58
V DG++DL H GH +L QA+ L+VG DE KG V +A ER E +
Sbjct 37 VYADGIYDLFHFGHARSLEQAKLAFPNNTYLLVGCCNDETTHKYKG-RTVMTAEERYESL 95
Query 59 ASCKWVDEVVVGTPYAVSTHLLDALSCEFAAHGDDWVCSST--GEDAYAGPRAAGRMKTF 116
CKWVDEV+ P+ V+ LD ++ AH S+ G+D Y + GR K
Sbjct 96 RHCKWVDEVIPDAPWVVNQEFLDKHQIDYVAHDSLPYADSSGAGKDVYEFVKKVGRFKET 155
Query 117 ARTE 120
RTE
Sbjct 156 QRTE 159
> At4g15130
Length=298
Score = 75.5 bits (184), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 46/126 (36%), Positives = 63/126 (50%), Gaps = 11/126 (8%)
Query 2 VQTDGVFDLLHSGHLNALRQARE--LGCCLVVGINGDEAVLAAKGFLPVYSAAERAEVIA 59
V DG+FDL H GH A+ QA++ L+VG DE KG V + +ER E +
Sbjct 23 VYADGIFDLFHFGHARAIEQAKKSFPNTYLLVGCCNDEITNKFKG-KTVMTESERYESLR 81
Query 60 SCKWVDEVVVGTPYAVSTHLLDALSCEFAAH-----GDDWVCSSTGEDAYAGPRAAGRMK 114
CKWVDEV+ P+ ++T LD ++ AH D S G D Y ++ G+ K
Sbjct 82 HCKWVDEVIPDAPWVLTTEFLDKHKIDYVAHDALPYAD---TSGAGNDVYEFVKSIGKFK 138
Query 115 TFARTE 120
RTE
Sbjct 139 ETKRTE 144
> YGR202c
Length=424
Score = 75.1 bits (183), Expect = 3e-14, Method: Composition-based stats.
Identities = 41/120 (34%), Positives = 60/120 (50%), Gaps = 3/120 (2%)
Query 2 VQTDGVFDLLHSGHLNALRQAREL--GCCLVVGINGDEAVLAAKGFLPVYSAAERAEVIA 59
+ DGVFDL H GH+ L Q ++ L+VG+ D+ KG L V + +R E +
Sbjct 106 IYADGVFDLFHLGHMKQLEQCKKAFPNVTLIVGVPSDKITHKLKG-LTVLTDKQRCETLT 164
Query 60 SCKWVDEVVVGTPYAVSTHLLDALSCEFAAHGDDWVCSSTGEDAYAGPRAAGRMKTFART 119
C+WVDEVV P+ V+ L ++ AH D S+ +D Y + G+ T RT
Sbjct 165 HCRWVDEVVPNAPWCVTPEFLLEHKIDYVAHDDIPYVSADSDDIYKPIKEMGKFLTTQRT 224
> ECU10g1590
Length=278
Score = 74.3 bits (181), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 40/118 (33%), Positives = 59/118 (50%), Gaps = 3/118 (2%)
Query 5 DGVFDLLHSGHLNALRQAREL--GCCLVVGINGDEAVLAAKGFLPVYSAAERAEVIASCK 62
DGV+DL H GH +L+QA+ L L+VG+ D+ + KG L V ERAE + C+
Sbjct 48 DGVYDLFHYGHARSLKQAKNLFPNVYLLVGVTDDDITVRLKGNL-VMDENERAEGLIHCR 106
Query 63 WVDEVVVGTPYAVSTHLLDALSCEFAAHGDDWVCSSTGEDAYAGPRAAGRMKTFARTE 120
+VDEV+ P+ ++ L +F AH D +D Y + G RT+
Sbjct 107 YVDEVITSAPWELTLEFLQKHKIDFVAHDDIPYKGEDKDDIYKFVKDMGMFIPIRRTK 164
> 7292098
Length=381
Score = 71.6 bits (174), Expect = 4e-13, Method: Composition-based stats.
Identities = 41/121 (33%), Positives = 60/121 (49%), Gaps = 3/121 (2%)
Query 2 VQTDGVFDLLHSGHLNALRQAREL--GCCLVVGINGDEAVLAAKGFLPVYSAAERAEVIA 59
V DG++DL H GH L QA+ + L+VG+ DE L KG V + ER E +
Sbjct 81 VYADGIYDLFHQGHARQLMQAKNVFPNVYLIVGVCNDELTLRMKG-RTVMNGFERYEAVR 139
Query 60 SCKWVDEVVVGTPYAVSTHLLDALSCEFAAHGDDWVCSSTGEDAYAGPRAAGRMKTFART 119
C++VDE+V P+ ++ ++ +F AH D + D YA +A G RT
Sbjct 140 HCRYVDEIVPNAPWTLNEEFIEEHKIDFVAHDDIPYGAGGVNDIYAPLKAKGMFVATERT 199
Query 120 E 120
E
Sbjct 200 E 200
> 7292096
Length=347
Score = 70.5 bits (171), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 41/121 (33%), Positives = 59/121 (48%), Gaps = 3/121 (2%)
Query 2 VQTDGVFDLLHSGHLNALRQAREL--GCCLVVGINGDEAVLAAKGFLPVYSAAERAEVIA 59
V DG++DL H GH L QA+ + L+VG+ DE KG V + ER E +
Sbjct 29 VYADGIYDLFHQGHARQLMQAKNIFPNVYLIVGVCNDELTHRMKG-RTVMNGFERYEGVR 87
Query 60 SCKWVDEVVVGTPYAVSTHLLDALSCEFAAHGDDWVCSSTGEDAYAGPRAAGRMKTFART 119
C++VDE+V P+ +S + +F AH D + +D YA +A G RT
Sbjct 88 HCRYVDEIVQNAPWTLSDEFIADNKIDFVAHDDIPYVTDGMDDIYAPLKARGMFVATERT 147
Query 120 E 120
E
Sbjct 148 E 148
> CE02591
Length=347
Score = 69.3 bits (168), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 41/121 (33%), Positives = 57/121 (47%), Gaps = 3/121 (2%)
Query 2 VQTDGVFDLLHSGHLNALRQAREL--GCCLVVGINGDEAVLAAKGFLPVYSAAERAEVIA 59
+ DG++DL H GH N LRQ +++ L+VG+ GD KG V S ER + +
Sbjct 68 IYADGIYDLFHHGHANQLRQVKKMFPNVYLIVGVCGDRDTHKYKG-RTVTSEEERYDGVR 126
Query 60 SCKWVDEVVVGTPYAVSTHLLDALSCEFAAHGDDWVCSSTGEDAYAGPRAAGRMKTFART 119
C++VDEV P+ + L L +F AH + ED Y R G RT
Sbjct 127 HCRYVDEVYREAPWFCTVEFLKNLKVDFIAHDAIPYVAPGEEDLYEKFRREGMFLETERT 186
Query 120 E 120
E
Sbjct 187 E 187
> CE27104
Length=302
Score = 67.0 bits (162), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 36/83 (43%), Positives = 49/83 (59%), Gaps = 1/83 (1%)
Query 2 VQTDGVFDLLHSGHLNALRQARELGCCLVVGINGDEAVLAAKG-FLPVYSAAERAEVIAS 60
V G FDL H+GHL+ L A++LG L+VGI GD+ V KG PV + ER I+S
Sbjct 151 VYVSGAFDLFHAGHLSFLEAAKDLGDYLIVGIVGDDDVNEEKGTIFPVMNLLERTLNISS 210
Query 61 CKWVDEVVVGTPYAVSTHLLDAL 83
K VDEV VG P ++ ++ +
Sbjct 211 LKIVDEVFVGVPAVTNSKFVNLI 233
Score = 57.0 bits (136), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 27/69 (39%), Positives = 42/69 (60%), Gaps = 1/69 (1%)
Query 2 VQTDGVFDLLHSGHLNALRQARELGCCLVVGINGDEAVLAAKGFLPVYSAAERAEVIASC 61
V TDG FD +H + L A++ G L+VGI+ D+ L G LP+++ ER +I++
Sbjct 20 VYTDGCFDFVHFANARLLWPAKQYGKKLIVGIHSDDE-LDNNGILPIFTDEERYRLISAI 78
Query 62 KWVDEVVVG 70
+WVDE + G
Sbjct 79 RWVDEEIAG 87
> CE19415
Length=183
Score = 63.2 bits (152), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 40/121 (33%), Positives = 56/121 (46%), Gaps = 21/121 (17%)
Query 2 VQTDGVFDLLHSGHLNALRQAREL--GCCLVVGINGDEAVLAAKGFLPVYSAAERAEVIA 59
V DG++D+ H GH L Q +++ L+VG+ DE L KG V S ER E +
Sbjct 44 VYADGIYDMFHYGHAKQLLQIKQMFPMVYLIVGVCSDENTLKFKG-PTVQSENERYESVR 102
Query 60 SCKWVDEVVVGTPYAVSTHLLDALSCEFAAHGDDWVCSSTGEDAYAGPRAAGRMKTFART 119
C++VDEV+ P+ VS L+ + YA +AAGR RT
Sbjct 103 QCRYVDEVLKDAPWEVSVDFLEEMKM------------------YAEHKAAGRFLETTRT 144
Query 120 E 120
E
Sbjct 145 E 145
> At5g23330
Length=354
Score = 32.3 bits (72), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 35/123 (28%), Positives = 49/123 (39%), Gaps = 16/123 (13%)
Query 6 GVFDLLHSGHLNALRQARELGCCLVVGINGDEAVLAAKGFLPVYSAAERAEVIAS----C 61
G FD LH GH QA +G ++ G VL K P+ + +R V++S C
Sbjct 90 GKFDALHIGHRELAIQAARIGTPYLLSFVGLAEVLGWKPRAPIVAKCDRKRVLSSWASYC 149
Query 62 KWVDEVVVGTPYAVSTHL-----LDALSCEFAAHGDDWVCSST-GEDAYAGPRAAGRMKT 115
+ V +A HL ++ LS E VC GE+ G RA+G
Sbjct 150 GNIAPVEFEIEFASVRHLNPQQFVEKLSRELR------VCGVVAGENYRFGYRASGDASE 203
Query 116 FAR 118
R
Sbjct 204 LVR 206
> CE28145_1
Length=246
Score = 30.4 bits (67), Expect = 0.84, Method: Compositional matrix adjust.
Identities = 15/41 (36%), Positives = 22/41 (53%), Gaps = 1/41 (2%)
Query 6 GVFDLLHSGHLNALRQARELGC-CLVVGINGDEAVLAAKGF 45
G FD LH+GH L +A EL +VVG+ + ++ F
Sbjct 106 GTFDRLHNGHKVLLNKAAELASDVIVVGVTDKDMIIKKSLF 146
> At5g08340
Length=508
Score = 30.0 bits (66), Expect = 1.0, Method: Composition-based stats.
Identities = 17/55 (30%), Positives = 25/55 (45%), Gaps = 0/55 (0%)
Query 6 GVFDLLHSGHLNALRQARELGCCLVVGINGDEAVLAAKGFLPVYSAAERAEVIAS 60
G FD LH GH QA +G ++ G VL + P+ + +R V+ S
Sbjct 254 GKFDALHIGHRELTIQASRIGAPYLLSFVGMAEVLGWEPRAPIVAKCDRQRVLTS 308
> SPAC29A4.02c_2
Length=156
Score = 29.6 bits (65), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 25/49 (51%), Gaps = 7/49 (14%)
Query 11 LHSGHLNALRQARE--LGCCLVVGINGDEAVLAA---KG--FLPVYSAA 52
L G L +R+ GCC+V+G NGD + A KG ++P + A
Sbjct 64 LIGGFFQRLEASRKYIFGCCVVIGENGDNTITGAFVIKGHDYVPAFDVA 112
> ECU06g1030
Length=341
Score = 28.9 bits (63), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 15/39 (38%), Positives = 19/39 (48%), Gaps = 9/39 (23%)
Query 9 DLLHSGHLNALRQARELGCCLVVGINGDEAVLAAKGFLP 47
DL+ H N LR+ GING E V+ +GF P
Sbjct 116 DLIRIAHFNVLRR---------FGINGCEIVVRKRGFAP 145
> Hs4557597
Length=2705
Score = 28.1 bits (61), Expect = 4.6, Method: Composition-based stats.
Identities = 11/27 (40%), Positives = 16/27 (59%), Gaps = 0/27 (0%)
Query 49 YSAAERAEVIASCKWVDEVVVGTPYAV 75
Y+ E+ + I KW DE V G+P+ V
Sbjct 2676 YTVKEKGDYILIVKWGDESVPGSPFKV 2702
> 7300550
Length=438
Score = 27.3 bits (59), Expect = 6.2, Method: Composition-based stats.
Identities = 16/45 (35%), Positives = 26/45 (57%), Gaps = 2/45 (4%)
Query 48 VYSAAERAEVIASCKWVDEVVVGTPYAVSTHLLDALSCEFAAHGD 92
+Y+ A+RA I+ ++ V + T Y +S HL+D + F A GD
Sbjct 330 MYTLADRA--ISKDEFSRAVKICTGYKLSPHLIDTVFAIFDADGD 372
> At2g18250
Length=176
Score = 26.9 bits (58), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 20/46 (43%), Positives = 24/46 (52%), Gaps = 2/46 (4%)
Query 1 AVQTDGVFDLLHSGHLNALRQARELGC-CLVVGINGDEAVLAAKGF 45
AV G FD LH GH L+ A EL +VVG+ D +L K F
Sbjct 17 AVVLGGTFDRLHDGHRMFLKAAAELARDRIVVGV-CDGPMLTKKQF 61
Lambda K H
0.320 0.134 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1160968786
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40