bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1633_orf1
Length=133
Score E
Sequences producing significant alignments: (Bits) Value
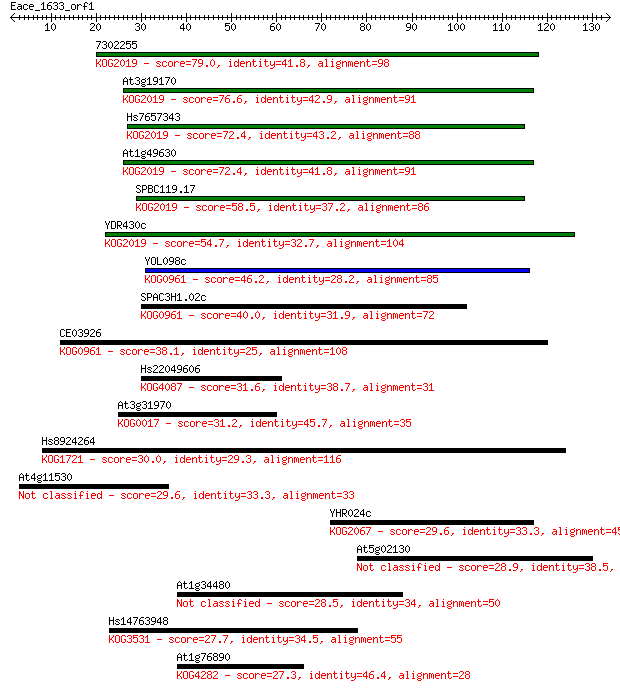
7302255 79.0 2e-15
At3g19170 76.6 1e-14
Hs7657343 72.4 2e-13
At1g49630 72.4 2e-13
SPBC119.17 58.5 3e-09
YDR430c 54.7 5e-08
YOL098c 46.2 2e-05
SPAC3H1.02c 40.0 0.001
CE03926 38.1 0.005
Hs22049606 31.6 0.42
At3g31970 31.2 0.59
Hs8924264 30.0 1.4
At4g11530 29.6 1.8
YHR024c 29.6 1.8
At5g02130 28.9 2.7
At1g34480 28.5 4.1
Hs14763948 27.7 6.4
At1g76890 27.3 7.4
> 7302255
Length=1112
Score = 79.0 bits (193), Expect = 2e-15, Method: Composition-based stats.
Identities = 41/99 (41%), Positives = 60/99 (60%), Gaps = 2/99 (2%)
Query 20 VYPEACELKYAGIVFSEMKGVFSSPELLEEQRRFQELFPDVLTYREVAGGDPEAIPQLTI 79
++ + +L G+V++EMKG FS + Q +FPD TYR V+GG+P IP+L
Sbjct 213 IFDKQSKLVIKGVVYNEMKGAFSENAQVFSQNLLNNIFPD-HTYRHVSGGNPLEIPKLAY 271
Query 80 PHLADFHKKYYAPSNAWVVLYGPDPVQERLSLID-DYLT 117
L +FHKKYY PSNA + YG + L+L+D +YL+
Sbjct 272 NDLVEFHKKYYHPSNARIYSYGLFDASKTLALLDEEYLS 310
> At3g19170
Length=1052
Score = 76.6 bits (187), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 39/91 (42%), Positives = 57/91 (62%), Gaps = 1/91 (1%)
Query 26 ELKYAGIVFSEMKGVFSSPELLEEQRRFQELFPDVLTYREVAGGDPEAIPQLTIPHLADF 85
++ Y G+VF+EMKGV+S P+ + + Q L P+ TY +GGDP+ IP LT +F
Sbjct 224 DISYKGVVFNEMKGVYSQPDNILGRIAQQALSPEN-TYGVDSGGDPKDIPNLTFEEFKEF 282
Query 86 HKKYYAPSNAWVVLYGPDPVQERLSLIDDYL 116
H++YY PSNA + YG D RL ++ +YL
Sbjct 283 HRQYYHPSNARIWFYGDDDPVHRLRVLSEYL 313
> Hs7657343
Length=1038
Score = 72.4 bits (176), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 38/88 (43%), Positives = 50/88 (56%), Gaps = 1/88 (1%)
Query 27 LKYAGIVFSEMKGVFSSPELLEEQRRFQELFPDVLTYREVAGGDPEAIPQLTIPHLADFH 86
L + G+VF+EMKG F+ E + Q L PD TY V+GGDP IP+LT L FH
Sbjct 196 LVFKGVVFNEMKGAFTDNERIFSQHLQNRLLPD-HTYSVVSGGDPLCIPELTWEQLKQFH 254
Query 87 KKYYAPSNAWVVLYGPDPVQERLSLIDD 114
+Y PSNA YG P+++ L I +
Sbjct 255 ATHYHPSNARFFTYGNFPLEQHLKQIHE 282
> At1g49630
Length=1076
Score = 72.4 bits (176), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 38/91 (41%), Positives = 57/91 (62%), Gaps = 5/91 (5%)
Query 26 ELKYAGIVFSEMKGVFSSPELLEEQRRFQELFPDVLTYREVAGGDPEAIPQLTIPHLADF 85
++ Y G+VF+EMKGV+S P+ + + L P+ TY +GGDP+ IP+LT +F
Sbjct 251 DISYKGVVFNEMKGVYSQPDNILGR----ALCPEN-TYGVDSGGDPKDIPKLTFEKFKEF 305
Query 86 HKKYYAPSNAWVVLYGPDPVQERLSLIDDYL 116
H++YY PSNA + YG D RL ++ +YL
Sbjct 306 HRQYYHPSNARIWFYGDDDPVHRLRVLSEYL 336
> SPBC119.17
Length=882
Score = 58.5 bits (140), Expect = 3e-09, Method: Composition-based stats.
Identities = 32/86 (37%), Positives = 47/86 (54%), Gaps = 1/86 (1%)
Query 29 YAGIVFSEMKGVFSSPELLEEQRRFQELFPDVLTYREVAGGDPEAIPQLTIPHLADFHKK 88
+ G+V++EMKG S + Q LF Y +GGDP AIP L L FH+
Sbjct 185 FNGVVYNEMKGQVSDSSYIFYMLFQQHLFQGT-AYGFNSGGDPLAIPDLKYEELVKFHRS 243
Query 89 YYAPSNAWVVLYGPDPVQERLSLIDD 114
+Y PSNA ++ YG P+++ LS + +
Sbjct 244 HYHPSNAKILSYGSFPLEDNLSALSE 269
> YDR430c
Length=989
Score = 54.7 bits (130), Expect = 5e-08, Method: Composition-based stats.
Identities = 34/104 (32%), Positives = 53/104 (50%), Gaps = 5/104 (4%)
Query 22 PEACELKYAGIVFSEMKGVFSSPELLEEQRRFQELFPDVLTYREVAGGDPEAIPQLTIPH 81
PE+ + + G+V++EMKG S+ + Q ++P + +GGDP I L
Sbjct 172 PES-NIVFKGVVYNEMKGQISNANYYFWSKFQQSIYPSL----NNSGGDPMKITDLRYGD 226
Query 82 LADFHKKYYAPSNAWVVLYGPDPVQERLSLIDDYLTRHGLGAYK 125
L DFH K Y PSNA YG P+ + L +++ + +G A K
Sbjct 227 LLDFHHKNYHPSNAKTFTYGNLPLVDTLKQLNEQFSGYGKRARK 270
> YOL098c
Length=1037
Score = 46.2 bits (108), Expect = 2e-05, Method: Composition-based stats.
Identities = 24/85 (28%), Positives = 45/85 (52%), Gaps = 0/85 (0%)
Query 31 GIVFSEMKGVFSSPELLEEQRRFQELFPDVLTYREVAGGDPEAIPQLTIPHLADFHKKYY 90
G+VFSEM+ + + + + + +FP+ YR GG + + LT + FHK Y
Sbjct 148 GVVFSEMEAIETQGWYISGLEKQRLMFPEGSGYRSETGGLTKNLRTLTNDEIRQFHKSLY 207
Query 91 APSNAWVVLYGPDPVQERLSLIDDY 115
+ N V++ G P E L++++++
Sbjct 208 SSDNLCVIVCGNVPTDELLTVMEEW 232
> SPAC3H1.02c
Length=1036
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 23/74 (31%), Positives = 40/74 (54%), Gaps = 4/74 (5%)
Query 30 AGIVFSEMKGVFSSPE--LLEEQRRFQELFPDVLTYREVAGGDPEAIPQLTIPHLADFHK 87
+G+V+SEM+ SS + + R Q +P Y GG P + +L+I + ++HK
Sbjct 147 SGVVYSEMQNTQSSETDVMFDCMRTSQ--YPVTSGYYYETGGHPSELRKLSIEKIREYHK 204
Query 88 KYYAPSNAWVVLYG 101
+ Y PSN +++ G
Sbjct 205 EMYVPSNICLIVTG 218
> CE03926
Length=995
Score = 38.1 bits (87), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 27/109 (24%), Positives = 54/109 (49%), Gaps = 1/109 (0%)
Query 12 SPSEVDCKVYPEACELKYAGIVFSEMKGVFSSPELLEEQRRFQELFPDVLTYREVAGGDP 71
+ S+ +V+ E AG+V+SEM+ S E + +++ + ++P Y GG
Sbjct 129 TASQFATEVHHITGEGNDAGVVYSEMQDHESEMESIMDRKTKEVIYPPFNPYAVDTGGRL 188
Query 72 EAIPQ-LTIPHLADFHKKYYAPSNAWVVLYGPDPVQERLSLIDDYLTRH 119
+ + + T+ + D+HKK+Y SN V + G + L ++++ H
Sbjct 189 KNLRESCTLEKVRDYHKKFYHLSNMVVTVCGMVDHDQVLEIMNNVENEH 237
> Hs22049606
Length=828
Score = 31.6 bits (70), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 12/31 (38%), Positives = 19/31 (61%), Gaps = 0/31 (0%)
Query 30 AGIVFSEMKGVFSSPELLEEQRRFQELFPDV 60
AG+ +M G+F P +L QRR Q+ P++
Sbjct 193 AGLNTVKMCGIFGGPRMLHTQRRLQDKSPNL 223
> At3g31970
Length=1329
Score = 31.2 bits (69), Expect = 0.59, Method: Compositional matrix adjust.
Identities = 16/38 (42%), Positives = 22/38 (57%), Gaps = 3/38 (7%)
Query 25 CELKYAGIVFSE---MKGVFSSPELLEEQRRFQELFPD 59
C L G VF++ +K +F+ PEL QRR+ EL D
Sbjct 760 CVLMQRGKVFTDHKSLKYIFTQPELNLRQRRWMELVAD 797
> Hs8924264
Length=754
Score = 30.0 bits (66), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 34/119 (28%), Positives = 52/119 (43%), Gaps = 19/119 (15%)
Query 8 GSGSSPSEVDCKVYPEACE---LKYAGIVFSEMKGVFSSPELLEEQRRFQELFPDVLTYR 64
G GSS K YP CE L + + + EM G + L E R+ L P+V T
Sbjct 37 GQGSSLQ----KNYPPVCEIFRLHFRQLCYHEMSGPQEALSRLRELCRWW-LMPEVHT-- 89
Query 65 EVAGGDPEAIPQLTIPHLADFHKKYYAPSNAWVVLYGPDPVQERLSLIDDYLTRHGLGA 123
E I +L + L F WV L+ P+ +E +++++D+ RH G+
Sbjct 90 ------KEQILELLV--LEQFLSILPGELRTWVQLHHPESGEEAVAVVEDF-QRHLSGS 139
> At4g11530
Length=931
Score = 29.6 bits (65), Expect = 1.8, Method: Composition-based stats.
Identities = 11/33 (33%), Positives = 20/33 (60%), Gaps = 0/33 (0%)
Query 3 GSSPSGSGSSPSEVDCKVYPEACELKYAGIVFS 35
G+S S S P++ D +P+ C ++Y+ + FS
Sbjct 354 GASDKISESCPNKTDAYTWPDCCMVRYSNVSFS 386
> YHR024c
Length=482
Score = 29.6 bits (65), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 15/45 (33%), Positives = 23/45 (51%), Gaps = 1/45 (2%)
Query 72 EAIPQLTIPHLADFHKKYYAPSNAWVVLYGPDPVQERLSLIDDYL 116
E IP ++ +L D+ K+Y P N G P ++ L L + YL
Sbjct 177 ELIPSISKYYLLDYRNKFYTPENTVAAFVGV-PHEKALELTEKYL 220
> At5g02130
Length=420
Score = 28.9 bits (63), Expect = 2.7, Method: Composition-based stats.
Identities = 20/54 (37%), Positives = 24/54 (44%), Gaps = 2/54 (3%)
Query 78 TIPHLADFHKKYYAPSNAWVVLYGPDPVQERLS--LIDDYLTRHGLGAYKPPPV 129
T H D H K A ++YG QER S LI + L R L K PP+
Sbjct 290 TEEHYGDNHPKVGVILTAVALMYGNKAKQERSSSILIQEGLYRKALELMKAPPL 343
> At1g34480
Length=602
Score = 28.5 bits (62), Expect = 4.1, Method: Composition-based stats.
Identities = 17/50 (34%), Positives = 25/50 (50%), Gaps = 6/50 (12%)
Query 38 KGVFSSPELLEEQRRFQELFPDVLTYREVAGGDPEAIPQLTIPHLADFHK 87
K V+ EL EE E++ D+L ++E+AGG I + H HK
Sbjct 281 KDVWDGKELEEEP---DEIYKDILPFKEIAGG---IIQHFSHKHQMRLHK 324
> Hs14763948
Length=594
Score = 27.7 bits (60), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 19/55 (34%), Positives = 26/55 (47%), Gaps = 2/55 (3%)
Query 23 EACELKYAGIVFSEMKGVFSSPELLEEQRRFQELFPDVLTYREVAGGDPEAIPQL 77
E L Y G + + GV +S +LE +RR L +V E+ PEA P L
Sbjct 232 EDLRLAYGGGYYQNVNGVHASEPVLESRRRNSAL--EVTFATELEHSKPEADPTL 284
> At1g76890
Length=575
Score = 27.3 bits (59), Expect = 7.4, Method: Composition-based stats.
Identities = 13/30 (43%), Positives = 21/30 (70%), Gaps = 2/30 (6%)
Query 38 KGVFS--SPELLEEQRRFQELFPDVLTYRE 65
KG+F+ + EL+E+Q + Q+ F + L YRE
Sbjct 262 KGLFTKLTKELMEKQEKMQKRFLETLEYRE 291
Lambda K H
0.315 0.137 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1356426142
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40