bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1653_orf1
Length=136
Score E
Sequences producing significant alignments: (Bits) Value
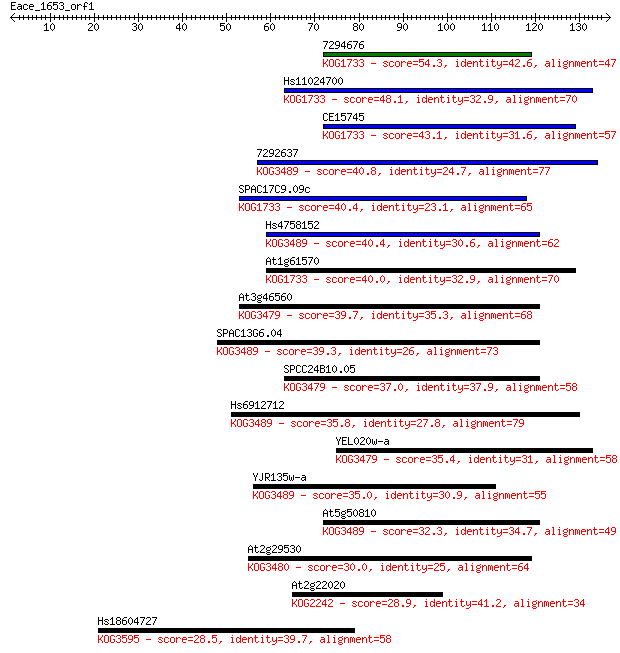
7294676 54.3 6e-08
Hs11024700 48.1 4e-06
CE15745 43.1 1e-04
7292637 40.8 7e-04
SPAC17C9.09c 40.4 0.001
Hs4758152 40.4 0.001
At1g61570 40.0 0.001
At3g46560 39.7 0.002
SPAC13G6.04 39.3 0.002
SPCC24B10.05 37.0 0.010
Hs6912712 35.8 0.026
YEL020w-a 35.4 0.031
YJR135w-a 35.0 0.038
At5g50810 32.3 0.24
At2g29530 30.0 1.3
At2g22020 28.9 2.9
Hs18604727 28.5 3.6
> 7294676
Length=92
Score = 54.3 bits (129), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 20/47 (42%), Positives = 33/47 (70%), Gaps = 0/47 (0%)
Query 72 AQSMLASLTSRCFTRCVDKPGRSLSSSQQQCLWQCAQRWSEVQHFVG 118
AQ ML+ +T +CF +C+ KPG+SL S++Q+C+ QC R+ + + V
Sbjct 24 AQEMLSKMTEKCFKKCIQKPGKSLDSTEQRCISQCMDRFMDAWNLVS 70
> Hs11024700
Length=95
Score = 48.1 bits (113), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 42/71 (59%), Gaps = 1/71 (1%)
Query 63 QLGLQQAL-EAQSMLASLTSRCFTRCVDKPGRSLSSSQQQCLWQCAQRWSEVQHFVGMRS 121
Q+ +Q A+ AQ +L +T +CF +C+ KPG SL +S+Q+C+ C R+ + + V
Sbjct 25 QVKVQIAVANAQELLQRMTDKCFRKCIGKPGGSLDNSEQKCIAMCMDRYMDAWNTVSRAY 84
Query 122 RALLQQQGGGL 132
+ LQ++ +
Sbjct 85 NSRLQRERANM 95
> CE15745
Length=108
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 18/57 (31%), Positives = 35/57 (61%), Gaps = 0/57 (0%)
Query 72 AQSMLASLTSRCFTRCVDKPGRSLSSSQQQCLWQCAQRWSEVQHFVGMRSRALLQQQ 128
AQ+++ ++ +C +C+ PG SL+S ++QCL +C R+ E + V + LQ++
Sbjct 34 AQNLVTDISEKCTNKCITAPGSSLASGEKQCLQRCMDRFMESWNLVSQTLQKRLQEE 90
> 7292637
Length=88
Score = 40.8 bits (94), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 19/77 (24%), Positives = 36/77 (46%), Gaps = 0/77 (0%)
Query 57 QQLAALQLGLQQALEAQSMLASLTSRCFTRCVDKPGRSLSSSQQQCLWQCAQRWSEVQHF 116
++L L +Q + + + C+ +C+ KP L + + CL C R+ +
Sbjct 12 KELQEFLLIEKQKAQVNAQIHEFNEICWEKCIGKPSTKLDHATETCLSNCVDRFIDTSLL 71
Query 117 VGMRSRALLQQQGGGLL 133
+ R +LQ++GGG L
Sbjct 72 ITQRFAQMLQKRGGGDL 88
> SPAC17C9.09c
Length=95
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 15/65 (23%), Positives = 35/65 (53%), Gaps = 8/65 (12%)
Query 53 QQQQQQLAALQLGLQQALEAQSMLASLTSRCFTRCVDKPGRSLSSSQQQCLWQCAQRWSE 112
+Q +Q+LA Q G +++ + CF +C+ +PG + +++ C+ +C +R+ +
Sbjct 23 KQIRQELAVAQAG--------ELISKINENCFDKCIPEPGSTFDPNEKSCVSKCMERYMD 74
Query 113 VQHFV 117
+ V
Sbjct 75 AWNIV 79
> Hs4758152
Length=97
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 19/69 (27%), Positives = 34/69 (49%), Gaps = 7/69 (10%)
Query 59 LAALQLGLQQALEAQS-------MLASLTSRCFTRCVDKPGRSLSSSQQQCLWQCAQRWS 111
L A+ LQ +E ++ ++ +T C+ +C+DKPG L S + C C +R+
Sbjct 12 LGAVDPQLQHFIEVETQKQRFQQLVHQMTELCWEKCMDKPGPKLDSRAEACFVNCVERFI 71
Query 112 EVQHFVGMR 120
+ F+ R
Sbjct 72 DTSQFILNR 80
> At1g61570
Length=87
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 23/70 (32%), Positives = 42/70 (60%), Gaps = 4/70 (5%)
Query 59 LAALQLGLQQALEAQSMLASLTSRCFTRCVDKPGRSLSSSQQQCLWQCAQRWSEVQHFVG 118
+ +++ L QA A+ ++ +L ++CF +CV KPG SL S+ C+ +C +R+ E +
Sbjct 22 MESVKTQLAQAY-AEELIETLRTKCFDKCVTKPGSSLGGSESSCISRCVERYMEATAII- 79
Query 119 MRSRALLQQQ 128
SR+L Q+
Sbjct 80 --SRSLFTQR 87
> At3g46560
Length=93
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 41/71 (57%), Gaps = 6/71 (8%)
Query 53 QQQQQQLAAL--QLGLQQALEAQSMLASLTSRCFTRCVDKPGR-SLSSSQQQCLWQCAQR 109
++ + ++A++ QL L+ +L M SL RCF CVD R SL ++ C+ +CA++
Sbjct 14 EEDKAKMASMIDQLQLRDSLR---MYNSLVERCFVDCVDSFTRKSLQKQEETCVMRCAEK 70
Query 110 WSEVQHFVGMR 120
+ + VGMR
Sbjct 71 FLKHTMRVGMR 81
> SPAC13G6.04
Length=98
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 19/73 (26%), Positives = 36/73 (49%), Gaps = 0/73 (0%)
Query 48 MTDDEQQQQQQLAALQLGLQQALEAQSMLASLTSRCFTRCVDKPGRSLSSSQQQCLWQCA 107
+ D + +Q +L+ QQ ++ Q + TS C+ +C+ G L S++QCL C
Sbjct 9 IADLSESEQLELSKFIESEQQKVKLQQAIHQFTSTCWPKCIGNIGNKLDKSEEQCLQNCV 68
Query 108 QRWSEVQHFVGMR 120
+R+ + + R
Sbjct 69 ERFLDCNFHIIKR 81
> SPCC24B10.05
Length=84
Score = 37.0 bits (84), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 22/65 (33%), Positives = 35/65 (53%), Gaps = 7/65 (10%)
Query 63 QLGLQQALEAQ------SMLASLTSRCFTRCV-DKPGRSLSSSQQQCLWQCAQRWSEVQH 115
Q L Q LEA+ +M ++LT CF+ CV D LS+ + +C+ +CA ++ +
Sbjct 9 QEHLTQVLEAKQLKEYLNMYSTLTQNCFSDCVQDFTSSKLSNKESECIAKCADKFLKHSE 68
Query 116 FVGMR 120
VG R
Sbjct 69 RVGQR 73
> Hs6912712
Length=83
Score = 35.8 bits (81), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 22/79 (27%), Positives = 38/79 (48%), Gaps = 4/79 (5%)
Query 51 DEQQQQQQLAALQLGLQQALEAQSMLASLTSRCFTRCVDKPGRSLSSSQQQCLWQCAQRW 110
DE + Q+ +AA Q Q + + + C+ +CV+KPG L S + CL C R+
Sbjct 8 DEAELQRLVAAEQ----QKAQFTAQVHHFMELCWDKCVEKPGNRLDSRTENCLSSCVDRF 63
Query 111 SEVQHFVGMRSRALLQQQG 129
+ + R ++Q+ G
Sbjct 64 IDTTLAITSRFAQIVQKGG 82
> YEL020w-a
Length=87
Score = 35.4 bits (80), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 18/59 (30%), Positives = 32/59 (54%), Gaps = 5/59 (8%)
Query 75 MLASLTSRCFTRCV-DKPGRSLSSSQQQCLWQCAQRWSEVQHFVGMRSRALLQQQGGGL 132
+ ++L RCFT CV D L++ +Q C+ +C++++ + VG R Q+Q L
Sbjct 27 LYSNLVERCFTDCVNDFTTSKLTNKEQTCIMKCSEKFLKHSERVGQR----FQEQNAAL 81
> YJR135w-a
Length=87
Score = 35.0 bits (79), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 17/56 (30%), Positives = 29/56 (51%), Gaps = 1/56 (1%)
Query 56 QQQLAALQLGLQQALEAQSMLASLTSRCFTRCVDKPGRS-LSSSQQQCLWQCAQRW 110
++++A G + Q + T+ CF +CV+ S LSS ++QCL C R+
Sbjct 17 KKEIATFLEGENSKQKVQMSIHQFTNICFKKCVESVNDSNLSSQEEQCLSNCVNRF 72
> At5g50810
Length=77
Score = 32.3 bits (72), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 17/50 (34%), Positives = 28/50 (56%), Gaps = 1/50 (2%)
Query 72 AQSMLASLTSRCFTRCVDK-PGRSLSSSQQQCLWQCAQRWSEVQHFVGMR 120
M++ +TS C+ +C+ PG SSS+ CL CAQR+ ++ + R
Sbjct 24 VNEMVSKMTSVCWDKCITSAPGSKFSSSESSCLTHCAQRYMDMSMIIMKR 73
> At2g29530
Length=83
Score = 30.0 bits (66), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 16/66 (24%), Positives = 32/66 (48%), Gaps = 5/66 (7%)
Query 55 QQQQLAALQLGLQQALEAQSMLASLTSRCFTRCVDKPGR--SLSSSQQQCLWQCAQRWSE 112
++Q + Q ++ +E + L CF +CVDK + L+ + C+ +C ++ +
Sbjct 11 KEQAFSMAQTEMEYRVE---LFNKLAQTCFNKCVDKRYKEAELNMGENSCIDRCVSKYWQ 67
Query 113 VQHFVG 118
V VG
Sbjct 68 VNGMVG 73
> At2g22020
Length=795
Score = 28.9 bits (63), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 14/36 (38%), Positives = 21/36 (58%), Gaps = 2/36 (5%)
Query 65 GLQQALEAQSMLASLTS--RCFTRCVDKPGRSLSSS 98
G Q EA + + T RCF+R +D+P RSL+ +
Sbjct 275 GFQPLQEAPTRFSFATELLRCFSRLLDRPDRSLADT 310
> Hs18604727
Length=2504
Score = 28.5 bits (62), Expect = 3.6, Method: Composition-based stats.
Identities = 23/62 (37%), Positives = 30/62 (48%), Gaps = 5/62 (8%)
Query 21 NNNTRLSSSITSARADHWIFCLLRIVLMTDDEQQQQQQ----LAALQLGLQQALEAQSML 76
N RL++ R H FC +VLM D +QQQ+ L L+ GL +EAQ
Sbjct 690 NAAERLATENRKLRKWHTTFCEKVVVLMNIDLLRQQQRWKDGLQELRTGL-ATVEAQGFQ 748
Query 77 AS 78
AS
Sbjct 749 AS 750
Lambda K H
0.320 0.127 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1425342594
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40