bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1657_orf1
Length=147
Score E
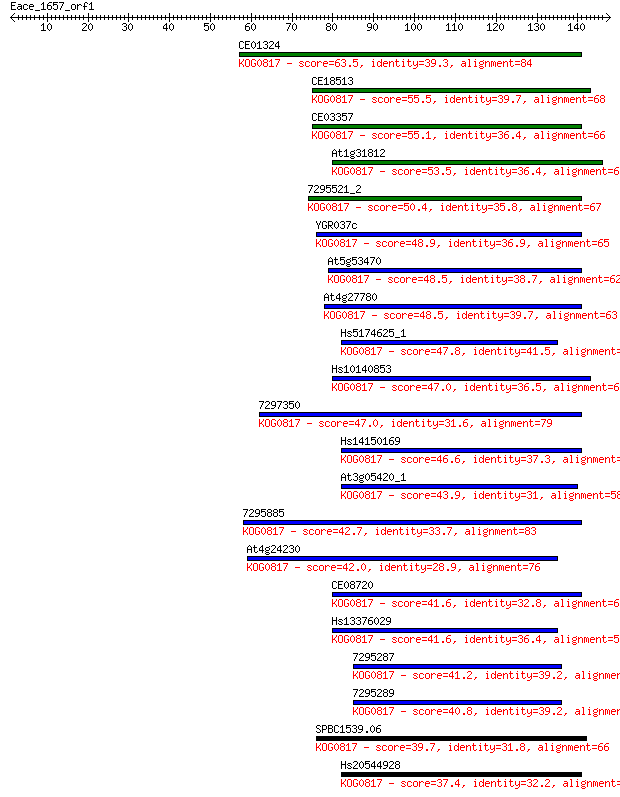
Sequences producing significant alignments: (Bits) Value
CE01324 63.5 2e-10
CE18513 55.5 4e-08
CE03357 55.1 5e-08
At1g31812 53.5 2e-07
7295521_2 50.4 1e-06
YGR037c 48.9 3e-06
At5g53470 48.5 4e-06
At4g27780 48.5 5e-06
Hs5174625_1 47.8 7e-06
Hs10140853 47.0 1e-05
7297350 47.0 1e-05
Hs14150169 46.6 2e-05
At3g05420_1 43.9 1e-04
7295885 42.7 3e-04
At4g24230 42.0 4e-04
CE08720 41.6 5e-04
Hs13376029 41.6 6e-04
7295287 41.2 6e-04
7295289 40.8 0.001
SPBC1539.06 39.7 0.002
Hs20544928 37.4 0.010
> CE01324
Length=146
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 33/86 (38%), Positives = 48/86 (55%), Gaps = 2/86 (2%)
Query 57 KALDEEFKG--YLRAAEQHKGPNPLSVEQQLAFYGLYKRATAGLCPSKQPSRFNIREYKK 114
K+LDE+F+ ++ A GP S+ QL Y LYK+AT+G C + QP F I + K
Sbjct 3 KSLDEQFEAAVWIINALPKNGPIKTSINDQLQMYSLYKQATSGKCDTIQPYFFQIEQRMK 62
Query 115 WEAWRACNSLSALEAKKEYVRRAREL 140
W AW ++ EAK +YV + +L
Sbjct 63 WNAWNQLGNMDEAEAKAQYVEKMLKL 88
> CE18513
Length=266
Score = 55.5 bits (132), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 27/72 (37%), Positives = 45/72 (62%), Gaps = 4/72 (5%)
Query 75 GPNPLSVEQQLAFYGLYKRATAGLCPSKQPSRFNIREYKKWEAWRACNSLSALEAKKEY- 133
GP S +++L FY L+K+AT G C +PS ++I+ KW AW ++++ EAK+ Y
Sbjct 18 GPIKTSTDEKLNFYALFKQATHGKCDLPKPSFYDIQGVYKWNAWNKLDNMTMDEAKQAYV 77
Query 134 ---VRRARELKK 142
V++ RE++K
Sbjct 78 DSIVQKIREVQK 89
> CE03357
Length=125
Score = 55.1 bits (131), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 24/66 (36%), Positives = 41/66 (62%), Gaps = 0/66 (0%)
Query 75 GPNPLSVEQQLAFYGLYKRATAGLCPSKQPSRFNIREYKKWEAWRACNSLSALEAKKEYV 134
GP S +Q+L FY L+K+A+ G + +P F+I E KKW++W+ +S EAK+ Y+
Sbjct 21 GPVATSNDQKLTFYSLFKQASIGDVNTDRPGIFSIIERKKWDSWKELEGVSQDEAKERYI 80
Query 135 RRAREL 140
+ ++
Sbjct 81 KALNDM 86
> At1g31812
Length=92
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 24/66 (36%), Positives = 40/66 (60%), Gaps = 0/66 (0%)
Query 80 SVEQQLAFYGLYKRATAGLCPSKQPSRFNIREYKKWEAWRACNSLSALEAKKEYVRRARE 139
S E L YGLYK+A G + +P F+++E KW+AW+A S+ EA +Y+ + ++
Sbjct 22 SNEDLLILYGLYKQAKFGPVDTSRPGMFSMKERAKWDAWKAVEGKSSEEAMNDYITKVKQ 81
Query 140 LKKLHA 145
L ++ A
Sbjct 82 LLEVAA 87
> 7295521_2
Length=231
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 24/67 (35%), Positives = 37/67 (55%), Gaps = 0/67 (0%)
Query 74 KGPNPLSVEQQLAFYGLYKRATAGLCPSKQPSRFNIREYKKWEAWRACNSLSALEAKKEY 133
K N + L FYG YK+AT G C + P ++ KW+AWR ++S A++ Y
Sbjct 10 KQSNSIGSADLLIFYGYYKQATNGPCKEQSPGLLQLQAKSKWQAWRNLGTMSQSAARQAY 69
Query 134 VRRAREL 140
V++ +EL
Sbjct 70 VQKLQEL 76
> YGR037c
Length=87
Score = 48.9 bits (115), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 24/65 (36%), Positives = 36/65 (55%), Gaps = 0/65 (0%)
Query 76 PNPLSVEQQLAFYGLYKRATAGLCPSKQPSRFNIREYKKWEAWRACNSLSALEAKKEYVR 135
P S ++ L Y LYK+AT G ++P FN+++ KWEAW S +A+KEY+
Sbjct 17 PTKPSTDELLELYALYKQATVGDNDKEKPGIFNMKDRYKWEAWENLKGKSQEDAEKEYIA 76
Query 136 RAREL 140
+L
Sbjct 77 LVDQL 81
> At5g53470
Length=338
Score = 48.5 bits (114), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 24/62 (38%), Positives = 35/62 (56%), Gaps = 0/62 (0%)
Query 79 LSVEQQLAFYGLYKRATAGLCPSKQPSRFNIREYKKWEAWRACNSLSALEAKKEYVRRAR 138
+S E QL YGLYK AT G C + QPS + KW+AW+ ++ EA ++Y+
Sbjct 117 VSNELQLQLYGLYKIATEGPCTAPQPSALKMTARAKWQAWQKLGAMPPEEAMEKYIDLVT 176
Query 139 EL 140
+L
Sbjct 177 QL 178
> At4g27780
Length=354
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 25/63 (39%), Positives = 36/63 (57%), Gaps = 2/63 (3%)
Query 78 PLSVEQQLAFYGLYKRATAGLCPSKQPSRFNIREYKKWEAWRACNSLSALEAKKEYVRRA 137
P V+QQL YGLYK AT G C + QPS + KW+AW+ ++ EA ++Y+
Sbjct 128 PSDVQQQL--YGLYKIATEGPCTAPQPSALKMTARAKWQAWQKLGAMPPEEAMEKYIEIV 185
Query 138 REL 140
+L
Sbjct 186 TQL 188
> Hs5174625_1
Length=98
Score = 47.8 bits (112), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 22/53 (41%), Positives = 30/53 (56%), Gaps = 0/53 (0%)
Query 82 EQQLAFYGLYKRATAGLCPSKQPSRFNIREYKKWEAWRACNSLSALEAKKEYV 134
E +L Y LYK+AT G C +P F++ KW+AW A SL A++ YV
Sbjct 25 EVKLKLYALYKQATEGPCNMPKPGVFDLINKAKWDAWNALGSLPKEAARQNYV 77
> Hs10140853
Length=104
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 23/63 (36%), Positives = 33/63 (52%), Gaps = 0/63 (0%)
Query 80 SVEQQLAFYGLYKRATAGLCPSKQPSRFNIREYKKWEAWRACNSLSALEAKKEYVRRARE 139
S E+ L YG YK+AT G +++P + KW+AW S +A K Y+ + E
Sbjct 38 SDEEMLFIYGHYKQATVGDINTERPGMLDFTGKAKWDAWNELKGTSKEDAMKAYINKVEE 97
Query 140 LKK 142
LKK
Sbjct 98 LKK 100
> 7297350
Length=90
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 25/80 (31%), Positives = 40/80 (50%), Gaps = 1/80 (1%)
Query 62 EFKGYLRAAEQHKGPNPLSVEQQLA-FYGLYKRATAGLCPSKQPSRFNIREYKKWEAWRA 120
E + + +AAE K N + L Y LYK+AT G C + +P + + KWEAW
Sbjct 3 ELQEFNQAAEDVKNLNTTPGDNDLLELYSLYKQATVGDCNTDKPGFLDFKGKAKWEAWNN 62
Query 121 CNSLSALEAKKEYVRRAREL 140
+S +A+ Y+ + + L
Sbjct 63 RKGMSNTDAQAAYITKVKAL 82
> Hs14150169
Length=282
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 22/59 (37%), Positives = 33/59 (55%), Gaps = 0/59 (0%)
Query 82 EQQLAFYGLYKRATAGLCPSKQPSRFNIREYKKWEAWRACNSLSALEAKKEYVRRAREL 140
EQ L Y YK+ G C + +PS F+ +KWEAW+A S +A +EY+ ++L
Sbjct 63 EQLLYLYARYKQVKVGNCNTPKPSFFDFEGKQKWEAWKALGDSSPSQAMQEYIAVVKKL 121
> At3g05420_1
Length=103
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 18/58 (31%), Positives = 34/58 (58%), Gaps = 0/58 (0%)
Query 82 EQQLAFYGLYKRATAGLCPSKQPSRFNIREYKKWEAWRACNSLSALEAKKEYVRRARE 139
+ L Y LY++AT G C + +PS + E KW++W+ ++ ++EA + +V+ E
Sbjct 42 DTALLLYALYQQATVGPCNTPKPSAWRPVEQSKWKSWQGLGTMPSIEAMRLFVKILEE 99
> 7295885
Length=324
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 28/86 (32%), Positives = 43/86 (50%), Gaps = 3/86 (3%)
Query 58 ALDEEFKGYLRAAE--QHKGPNPLSVEQQLAFYGLYKRATAG-LCPSKQPSRFNIREYKK 114
A++E F+ + + GP S L FYGL+K+AT G K+P ++I K
Sbjct 3 AIEERFQAAVNVIKGLPKNGPYQPSTSMMLKFYGLFKQATEGRPDVDKKPGFWDIVGKAK 62
Query 115 WEAWRACNSLSALEAKKEYVRRAREL 140
W+AW L+ EA + YV +E+
Sbjct 63 WQAWNDNRHLTKEEAMQRYVESLQEI 88
> At4g24230
Length=362
Score = 42.0 bits (97), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 22/76 (28%), Positives = 37/76 (48%), Gaps = 0/76 (0%)
Query 59 LDEEFKGYLRAAEQHKGPNPLSVEQQLAFYGLYKRATAGLCPSKQPSRFNIREYKKWEAW 118
L++ F + E+ + E ++ +GL+K AT G C QP I KW AW
Sbjct 231 LEKAFAAAVNLLEESGKAEEIGAEAKMELFGLHKIATEGSCREAQPMAVMISARAKWNAW 290
Query 119 RACNSLSALEAKKEYV 134
+ ++S EA ++Y+
Sbjct 291 QKLGNMSQEEAMEQYL 306
> CE08720
Length=86
Score = 41.6 bits (96), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 20/61 (32%), Positives = 32/61 (52%), Gaps = 0/61 (0%)
Query 80 SVEQQLAFYGLYKRATAGLCPSKQPSRFNIREYKKWEAWRACNSLSALEAKKEYVRRARE 139
S ++ L Y L+K+ T G + +P F+++ KW AW L+ +A+K YV E
Sbjct 20 SNDELLKLYALFKQGTVGDNTTDKPGMFDLKGKAKWSAWDEKKGLAKDDAQKAYVALVEE 79
Query 140 L 140
L
Sbjct 80 L 80
> Hs13376029
Length=305
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 29/55 (52%), Gaps = 0/55 (0%)
Query 80 SVEQQLAFYGLYKRATAGLCPSKQPSRFNIREYKKWEAWRACNSLSALEAKKEYV 134
S E+ L FY YK+AT G C +P ++ KW+AW + +S EA Y+
Sbjct 35 SYEEMLRFYSYYKQATMGPCLVPRPGFWDPIGRYKWDAWNSLGKMSREEAMSAYI 89
> 7295287
Length=94
Score = 41.2 bits (95), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 20/51 (39%), Positives = 31/51 (60%), Gaps = 0/51 (0%)
Query 85 LAFYGLYKRATAGLCPSKQPSRFNIREYKKWEAWRACNSLSALEAKKEYVR 135
L FYGL+K+AT G ++P +++ K+EAW + LS AK+ YV+
Sbjct 34 LEFYGLFKQATVGDVNIEKPGALALKDKAKYEAWSSNKGLSKEAAKEAYVK 84
> 7295289
Length=84
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 20/51 (39%), Positives = 30/51 (58%), Gaps = 0/51 (0%)
Query 85 LAFYGLYKRATAGLCPSKQPSRFNIREYKKWEAWRACNSLSALEAKKEYVR 135
L FYGL+K+AT G +P ++++ +EAW A LS AK+ YV+
Sbjct 24 LEFYGLFKQATVGDVNIDKPGILDLKKKAMYEAWNAHKGLSKDAAKEAYVK 74
> SPBC1539.06
Length=87
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 21/66 (31%), Positives = 33/66 (50%), Gaps = 3/66 (4%)
Query 76 PNPLSVEQQLAFYGLYKRATAGLCPSKQPSRFNIREYKKWEAWRACNSLSALEAKKEYVR 135
PN ++ L Y L+K+AT G +++P +++ KW AW S +A EY+
Sbjct 19 PNS---DELLKLYALFKQATVGDNNTEKPGLLDLKGKFKWNAWEELKGKSKEDAASEYIS 75
Query 136 RARELK 141
ELK
Sbjct 76 FVDELK 81
> Hs20544928
Length=490
Score = 37.4 bits (85), Expect = 0.010, Method: Composition-based stats.
Identities = 19/59 (32%), Positives = 30/59 (50%), Gaps = 0/59 (0%)
Query 82 EQQLAFYGLYKRATAGLCPSKQPSRFNIREYKKWEAWRACNSLSALEAKKEYVRRAREL 140
E L FY YK+AT G C +P ++ KW+AW + ++ EA YV +++
Sbjct 33 EMMLKFYSFYKQATEGPCKLSRPGFWDPIGRYKWDAWSSLGDMTKEEAMIAYVEEMKKI 91
Lambda K H
0.313 0.125 0.356
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1785281974
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40